Difference between revisions of "Projects/Slicer3/2007 Project Week Support for electron microscopy"
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | |[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | ||
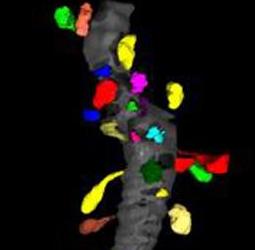
| − | |[[Image:Spines01.jpg|thumb|320px]] | + | |[[Image:Spines01.jpg|thumb|320px|A small fragment of a dendrite (gray) with segmented spines (color).]] |
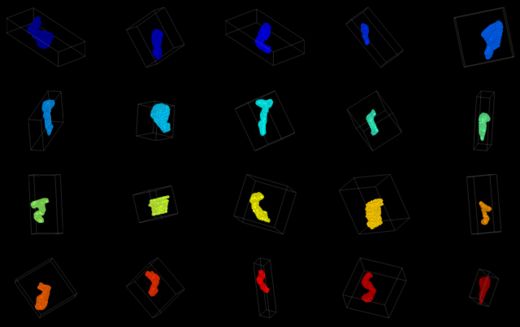
| − | |[[Image:SpineSample.jpg|thumb|520px]] | + | |[[Image:SpineSample.jpg|thumb|520px|A sample of spines plotted into individual axes.]] |
|} | |} | ||
Revision as of 21:46, 8 June 2007
Home < Projects < Slicer3 < 2007 Project Week Support for electron microscopy Return to Project Week Main Page |
Key Investigators
- UCSD/NCMIR: W. Bryan Smith, Mark Ellisman
- Isomics: Steve Pieper
Objective
Dendritic spines are the primary sites of excitatory neuronal communication in the brain. It is of fundamental interest to cellular neurobiologists to understand how the number, size, and shape of dendritic spines varies across brain regions, and as a function of experimental treatment within a given region. We would like to assess morphological properties of spines using Slicer.
Approach, Plan
Spines have been manually segmented from tilt-series electron tomograms. The spines need to be registered to a common coordinate system, and then a variety of measurements must be made on the size and shape of each spine in an effort to cluster the spines according to their morphologies.
The plan for the project week is to first try out 3D skeletonization filters in ITK to define the medial axis of each spine.
Progress
References
- Insert some refs here