Difference between revisions of "EM Segmentation For Orthopaedic Applications"
From NAMIC Wiki
| Line 9: | Line 9: | ||
*# Probability map information was created from a single subject used as the atlas image and filtered using a Gaussian filter. | *# Probability map information was created from a single subject used as the atlas image and filtered using a Gaussian filter. | ||
* Initial evaluation has been carried out on five subjects that also had been segmented using manual segmentation | * Initial evaluation has been carried out on five subjects that also had been segmented using manual segmentation | ||
| − | * Work is underway to develop a Slicer2.7 tutorial for this segmentation ([[Image: | + | * Work is underway to develop a Slicer2.7 tutorial for this segmentation ([[Image:Draft_EM_Segment_Tutorial_9_12.pdf|Draft Copy]]) |
'''To Do:''' | '''To Do:''' | ||
Revision as of 01:06, 30 October 2007
Home < EM Segmentation For Orthopaedic ApplicationsObjective:
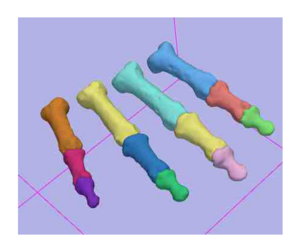
- To utilize the Slicer3 Expectation Maximization Algorithm for segmentation of the phalanx bones of the hand.
Progress:
- We have utilized the Slicer2.7 EM Segmentation Module for segmentation of the phalanx bones
- Registration was performed outside of SLicer using ITK registration algorithms that are available in the IaFeMesh software. This includes Thin plate spline, B-Spline and rigid registration algorithms.
- Probability map information was created from a single subject used as the atlas image and filtered using a Gaussian filter.
- Initial evaluation has been carried out on five subjects that also had been segmented using manual segmentation
- Work is underway to develop a Slicer2.7 tutorial for this segmentation (File:Draft EM Segment Tutorial 9 12.pdf)
To Do:
- Evaluate the Slicer3 EM Segmentation Module
- Update the the tutorial to support the Slicer3 Workflow
- Determine if registration tools in Slicer are adequate for this work.
Key Investigators:
- Iowa: Austin Ramme, Nicole Grosland, and Vincent Magnotta
Links:
Figures: