Difference between revisions of "Projects:ClusteringOfAnatomicallyDistinctFiberTracts"
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | Back to [[NA-MIC_Collaborations|NA-MIC_Collaborations]], [[Algorithm:MIT|MIT Algorithms]] | ||
| + | __TOC__ | ||
= Clustering Of Anatomically Distinct Fiber Tracts = | = Clustering Of Anatomically Distinct Fiber Tracts = | ||
| − | |||
| − | |||
| − | |||
| − | |||
Use fiber clustering in multiple subjects to create a model ("atlas") of tractography clusters. Perform automatic segmentation of novel tractography datasets (brains) by application of this atlas. Also transfer anatomical information via cluster labels in the atlas. | Use fiber clustering in multiple subjects to create a model ("atlas") of tractography clusters. Perform automatic segmentation of novel tractography datasets (brains) by application of this atlas. Also transfer anatomical information via cluster labels in the atlas. | ||
| − | + | = Description = | |
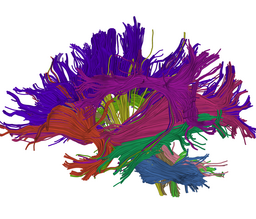
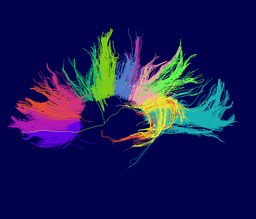
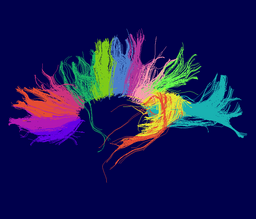
Atlas creation and automatic labeling has been performed in high-quality DTI datasets from Susumu Mori. Images showing example segmentation results are below. Work is underway to apply this atlas to segment additional datasets to define regions of interest that may be used in the study of schizophrenia. | Atlas creation and automatic labeling has been performed in high-quality DTI datasets from Susumu Mori. Images showing example segmentation results are below. Work is underway to apply this atlas to segment additional datasets to define regions of interest that may be used in the study of schizophrenia. | ||
| Line 21: | Line 19: | ||
[[Image:Th_corpus-SM_020001.png|[[Image:Th_corpus-SM_020001.png|Image:Th_corpus-SM_020001.png]]]] [[Image:Th_corpus-SM_140001.png|[[Image:Th_corpus-SM_140001.png|Image:Th_corpus-SM_140001.png]]]] [[Image:Th_corpus-SM_150001.png|[[Image:Th_corpus-SM_150001.png|Image:Th_corpus-SM_150001.png]]]] | [[Image:Th_corpus-SM_020001.png|[[Image:Th_corpus-SM_020001.png|Image:Th_corpus-SM_020001.png]]]] [[Image:Th_corpus-SM_140001.png|[[Image:Th_corpus-SM_140001.png|Image:Th_corpus-SM_140001.png]]]] [[Image:Th_corpus-SM_150001.png|[[Image:Th_corpus-SM_150001.png|Image:Th_corpus-SM_150001.png]]]] | ||
| + | = Key Investigators = | ||
| − | + | * MIT: Lauren O'Donnell | |
| − | + | * BWH: Carl-Fredrik Westin, Marek Kubicki, Martha Shenton | |
| − | * MIT | ||
| − | * BWH | ||
| − | |||
| − | |||
Latest revision as of 02:54, 27 November 2007
Home < Projects:ClusteringOfAnatomicallyDistinctFiberTractsBack to NA-MIC_Collaborations, MIT Algorithms
Clustering Of Anatomically Distinct Fiber Tracts
Use fiber clustering in multiple subjects to create a model ("atlas") of tractography clusters. Perform automatic segmentation of novel tractography datasets (brains) by application of this atlas. Also transfer anatomical information via cluster labels in the atlas.
Description
Atlas creation and automatic labeling has been performed in high-quality DTI datasets from Susumu Mori. Images showing example segmentation results are below. Work is underway to apply this atlas to segment additional datasets to define regions of interest that may be used in the study of schizophrenia.
Example Results
Selected anatomical regions, automatically labeled using the cluster atlas in 3 subjects.
Subdivisions of the corpus callosum, labeled using the cluster atlas in 3 subjects.
Key Investigators
- MIT: Lauren O'Donnell
- BWH: Carl-Fredrik Westin, Marek Kubicki, Martha Shenton