Difference between revisions of "2008 Winter Project Week:SmallAnimalEvalNCI"
(Initial edit - NCI Small Animal evaluation) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
| − | |[[Image: | + | |[[Image:Rt-flank-tumor-in-slicer-small.png|thumb|320px|Animal tumor in Slicer. Return to [[2008_Winter_Project_Week]] ]] |
| − | |valign="top"|[[Image: | + | |valign="top"|[[Image:small-animal-in-slicer.png |thumb|320px|Slicer Opening a Mouse Dataset]] |
|} | |} | ||
| Line 8: | Line 8: | ||
* KnowledgeVis: Curtis Lisle | * KnowledgeVis: Curtis Lisle | ||
* NCI ABCC: Jack Collins | * NCI ABCC: Jack Collins | ||
| − | * | + | * BWH: Kilian Pohl |
| − | |||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 31: | Line 30: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| + | ==Jan 2008 Project Week== | ||
| + | We are using a VisualSonics Vevo 770 ultrasound scanner. VisualSonics was helpful in showing us how to create an AVI movie of a 3D scan. Slices for the volume were extracted from the scan for a Slicer volume (see the picture on the left). | ||
| + | A very productive meeting with Kilian Pohl resulted in discovery that a tumor growth algorithm previously developed by Kllian and Sandy Wells will be working soon as a Slicer3 module. This module is a near perfect fit for the longitudinal study on small animal tumors underway at the NCI. Our project team will follow-up with Kilian during the next two quarters to test the module on our existing datasets. | ||
</div> | </div> | ||
| Line 38: | Line 40: | ||
</div> | </div> | ||
| − | |||
===References=== | ===References=== | ||
Latest revision as of 17:18, 6 February 2008
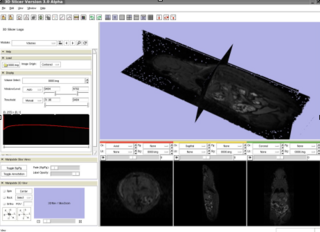
Home < 2008 Winter Project Week:SmallAnimalEvalNCI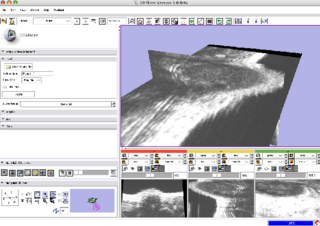 Animal tumor in Slicer. Return to 2008_Winter_Project_Week |
Key Investigators
- KnowledgeVis: Curtis Lisle
- NCI ABCC: Jack Collins
- BWH: Kilian Pohl
Objective
The NA-MIC toolset seems ready to be of substantial value in clinical research for Small Animal Imaging applications. The Advanced Biomedical Computing Center of the National Cancer Institute currently supports multiple Principal Investigators using many different medical imaging analysis programs. Greater efficiency and cooperation could result between Investigators when common tools are more widely used and supported by technical staff. The purpose of this project is to evaluate the NA-MIC toolset for installation at NCI ABCC and use by Small Animal Imaging researchers.
Approach, Plan
Our Approach for this project is to begin with small animal datasets in two different modalities: 3D Ultrasound and microPET. We will work to ingest these with Slicer3 and prepare the datasets by removing artifacts unique to these modalities. Finally, we will determine if any of the current segmentation algorithms underway are effective at identifying tumors and tumor change over time.
We will bring two datasets (3D ultrasound and microPET) to the AHM and work during the week to accomplish format injest and basic image processing and segmentation. After ingest is accomplished, Slicer3 will be installed at NCI ABCC and Small Animal Imaging Program researchers will be briefed on the results. If successful, a workflow will be installed for NCI researchers to upload, process, and disseminate their datasets.
Progress
Jan 2008 Project Week
We are using a VisualSonics Vevo 770 ultrasound scanner. VisualSonics was helpful in showing us how to create an AVI movie of a 3D scan. Slices for the volume were extracted from the scan for a Slicer volume (see the picture on the left).
A very productive meeting with Kilian Pohl resulted in discovery that a tumor growth algorithm previously developed by Kllian and Sandy Wells will be working soon as a Slicer3 module. This module is a near perfect fit for the longitudinal study on small animal tumors underway at the NCI. Our project team will follow-up with Kilian during the next two quarters to test the module on our existing datasets.