Difference between revisions of "MBIRN:BIRNDUP:Distribution"
| Line 66: | Line 66: | ||
cvs -d :pserver:anonymous@cvs.spl.harvard.edu:/projects/cvs/slicer checkout birndup | cvs -d :pserver:anonymous@cvs.spl.harvard.edu:/projects/cvs/slicer checkout birndup | ||
| − | A web interface to cvs is viewable here: http:// | + | A web interface to cvs is viewable here: |
| + | http://www.na-mic.org/ViewVC/index.cgi/birndup/?root=Slicer2 | ||
The user interface requires a current cvs version Slicer2. See the instructions [[Slicer:Slicer_2.6_Building|here ]]. | The user interface requires a current cvs version Slicer2. See the instructions [[Slicer:Slicer_2.6_Building|here ]]. | ||
Revision as of 12:55, 3 May 2008
Home < MBIRN:BIRNDUP:DistributionThis page includes information about the distribution of the BIRN Deidentification and Upload Pipeline (BIRNDUP).
Note: this software is experimental. Although it is functional as described, further development is on hold pending further feedback from the BIRN data sharing groups about the extent of the need for facial deidentification (if it is needed at all for MRI data).
Contents
Features
BIRNDUP supports:
- Removes patient identifiers from DICOM headers and replaces them with valid, but non-identifyable fields (Note: only these fields are removed.)
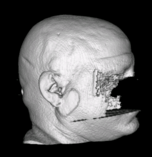
- Removes the faces from MR brain scans and replaces the data with zero valued voxels
- Sorts DICOM studies into the BIRN-standard directory hierarchy
- Provides an interactive review step to confirm that images have been correctly deidentified by showing 3D renderings of the defaced images
- Pseudorandom one-way hash BIRNID generated automatically from Subject ID and Site ID so that subsequent uploads always get the same BIRNID for longitudinal studies.
Supported Data Formats and Image Types
- Only DICOM data is supported
- A T1 scan is used for the defacing calculation. Each additional series in the study are masked by registering it to the T1.
Platform Requirements
Linux on x86. Red Hat Enterprise Linux tested, other Linux platforms probably also work.
Processing a single study can take approximately 2 hours and take about 200Meg of disk space. At least 1Gig of RAM is suggested.
The screen is used to render movies during part of the process. Please turn off any screen savers and do not use the display for any other purposes while the program is running.
Installation
Directory Hierarchy
Install the code in any convenient directory on your machine. Put slicer2 and the birndup distribution at the same level. By default, a directory called 'deface' is needed at the same level as a place to store your projects and linktable. The directory structure should look as follows:
% cd <install dir> % ls birndup deface slicer2 % ls birndup atlas bin dcanon fsl README scripts
The command line to start the BIRNDUP interface is:
<install dir>/slicer2/slicer2-linux-x86 -y --all-info --no-tkcon --eval package require BIRNDUP ,. dup_main | & tee /tmp/dataprovraw.txt
you may wish to create a handy alias or shell script wrapper for this command. No additional environment variables are needed (everything is found local to the install directory). For example:
alias birndup ~/birndup/slicer2/slicer2-linux-x86 -y --all-info --no-tkcon --eval package require BIRNDUP ,. dup_main
The final pipe to tee will record the raw unparsed data provenance information in a temporary text file. To convert it to xml, use this command line:
<install dir>/birndup/scripts/dataprov_progelem.tcl /tmp/dataprovraw.txt > /tmp/dataprov.xml
Packaged Distribution
Packaged distributions of this code will be released after further testing.
Development Versions
The distribution software for BIRNDUP can be obtained as follows
cvs -d :pserver:anonymous@cvs.spl.harvard.edu:/projects/cvs/slicer login
respond with bwhspl when prompted for password.
cvs -d :pserver:anonymous@cvs.spl.harvard.edu:/projects/cvs/slicer checkout birndup
A web interface to cvs is viewable here: http://www.na-mic.org/ViewVC/index.cgi/birndup/?root=Slicer2
The user interface requires a current cvs version Slicer2. See the instructions here .
Usage
Interactive Mode
Step by Step Instructions
This powerpoint presentation has an overview of the methodology plus step by step instructions.
Output Directory Format
Batch Mode for Defacing
See usage information here.
Batch Mode for Non-Defacing
To call the underlying code, you can use the command
dcanon
from the dcanon directory of the distribution (see cvs checkout information above).
Usage is:
usage: dcanon [options] <indir> [outdir] options are: -nostrip don't apply the skull stripping algorithm -noanon don't remove patient tags from header -watershed run mri_watershed to generate mask (skull stripping) -deface run mri_deface to generate mask (the default) -convert run mri_convert to generate mask (basically a no-op for testing) -mask <maskvol> run mri_mask to apply mask to <indir> -force delete outdir if it exists -version print out the version info for this script and exit -patname override the default patname of anon
For example, to remove identifiers from dicom files without doing any defacing or skull stripping, you can use something like:
dcanon -nostrip data/dicoms data/anonymized-dicoms
Output Directory Hierarchy
In the output directory (the DEFACE_DIR as configured in the Preferences Dialog) the working copies of all the image data processed by BIRNDUP. The format of the data is as follows:
- Each Study is stored in a unique directory with the following pattern:
<DEFACE_DIR>/Project_<Project>/<BIRNID>/Visit_<Visit>/Study_<Study>/Raw_Data
- <Project>, BIRNID, <Visit>, and <Study> are as specified in the Sort Pane
The Study Directory contains:
- 001, 002, ... <-- these are the original dicom image series (NOT deidentified!)
- 001-anon, 002-anon, ... <-- the deidentified dicom series to upload
- each <series>-anon directory contains a Deface subdirectory with png formated images of the volume rendered face files plus the axial, sagittal, and coronal images.
- upload_list.txt <-- the series have been approved for upload
- defer_list.txt <-- the series which should not be upload
- source_directory <-- text file with the original sort directory
- ready_for_upload <-- an empty file as a flag (if it exists, study is approved for upload)
- deidentify_operations <-- list of deidentification commands
Licensing
BIRNDUP is built using technology collected from mBIRN Partner sites and from 3rd party packages; as a result, different parts of the program are covered by different licenses.
- The User Interface is based on 3D Slicer: [1]
- The birndup/bin/mri_* binaries are part of FreeSurfer: [2] (note: only non-clinical, non-commercial uses are allowed).
- The flirt binary is part of FSL: [3] (note: only non-clinical, non-commercial uses are allowed).
- DICOM tools from [4] and [5]
- Other software in the distribution is covered by the BIRN Public License [6]
Acknowledgements
BIRNDUP is a joint work of the Morphometry BIRN (http://www.nbirn.net) which draws on technology developed by the collaborator sites including the Neuroimage Analysis Center (NAC at BWH) and the Martinos Center at MGH.