Difference between revisions of "NA-MIC NCBC Collaboration:Automated FE Mesh Development"
| Line 54: | Line 54: | ||
*Gassman EE, Powell SM, Kallemeyn NA, Devries NA, Shivanna KH, Magnotta VA, Ramme AJ, Adams BD, Grosland NM. [http://www.springerlink.com/content/b15376vwx00v5088/ Automated bony region identification using artificial neural networks: reliability and validation measurements]. Skeletal Radiol. 37(4):313-9, 2008. | *Gassman EE, Powell SM, Kallemeyn NA, Devries NA, Shivanna KH, Magnotta VA, Ramme AJ, Adams BD, Grosland NM. [http://www.springerlink.com/content/b15376vwx00v5088/ Automated bony region identification using artificial neural networks: reliability and validation measurements]. Skeletal Radiol. 37(4):313-9, 2008. | ||
*Grosland NM, Bafna R, Magnotta VA. [http://www.informaworld.com/smpp/content~db=all?content=10.1080/10255840802136143 Automated hexahedral meshing of anatomic structures using deformable registration]. Comput Methods Biomech Biomed Engin. 2008. | *Grosland NM, Bafna R, Magnotta VA. [http://www.informaworld.com/smpp/content~db=all?content=10.1080/10255840802136143 Automated hexahedral meshing of anatomic structures using deformable registration]. Comput Methods Biomech Biomed Engin. 2008. | ||
| − | *Magnotta, | + | *Magnotta V, Li W, Grosland N. [http://hdl.handle.net/1926/1490 Comparison of Displacement-Based and Force-Based Mapped Meshing]. Workshop on Computational Biomechanics for Medicine at MICCAI 2008. Insight Journal, http://hdl.handle.net/1926/1490 , 2008. |
==Meetings and Events Specific to this Collaboration== | ==Meetings and Events Specific to this Collaboration== | ||
Revision as of 00:37, 31 August 2008
Home < NA-MIC NCBC Collaboration:Automated FE Mesh DevelopmentBack to NA-MIC_External_Collaborations
Contents
Abstract
Musculoskeletal finite element (FE) analysis is an invaluable tool in orthopedic-related research. While it has provided significant biomechanical insight, the demands associated with modeling the geometrically complex structures of the human body often limit its utility. The often-prohibitive amount of model development time is further compounded by the time required to process medical image datasets to identify the distinct anatomical structures of interest. Yet this process is a necessary preprocessing step for model development. As a result, most of the analyses reported in the literature refer to 'average' bone geometry. The broad objective of our research plan is to integrate and expand methods to automate the development of specimen- / patient-specific finite element (FE) models into the NA-MIC toolkit. In pursuit of this objective we propose to merge unique technologies to automate image dataset segmentation; material property extraction and assignment; and direct FE model development (automated meshing). While direct physical scans of the bones of interest will be used to validate the automated image segmentation routines, experimental cadaveric contact stress measurements will provide a standard against which to validate the FE contact formulations. Furthermore, the FE models generated by our software package will be compared to models of the same bone(s) created via a commercial pre-processing package. While the bones/joints of the upper extremity represent the primary structures of interest proposed in this application, the tools will be applicable to many orthopedic applications. In addition to expanding the NA-MIC toolkit beyond the brain, the proposed project will expand the image segmentation routines and finite element meshing routines currently available. This proposal will ultimately yield specimen-specific FE models of the various joints of the upper extremity. Such models will position us to provide information about the load transfer, characteristics of the normal joints and in the future to demonstrate, for example, the effects of ligamentous instabilities, posttraumatic misalignments, fractures, and various surgical procedures.
Grant#
1R01EB005973
Key Personnel
Nicole Grosland, Vincent Magnotta, Steve Pieper, and Simon Warfield.
Projects
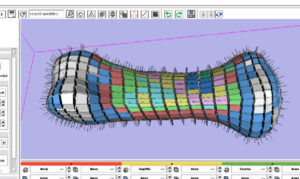
An initial version of the meshing tool was recently released as a standalone application called IA-FEMesh.
Active projects in this collaboration are:
Meshing Algorithms
- Voxel Meshing Module (Iowa)
- Novel Hexahedral Meshing Algorithms (Iowa)
- Mapped Hexahedral Meshing (Iowa)
- Hex vs Tet Mesh Comparisons (Iowa/Isomics/BWH)
Automated Segmentation
Image Registration
- Evaluation of Inter-Modality Registration (Iowa/Isomics)
Validation
Mesh Quality Visualization
- FE Mesh Quality Visualization (Iowa/Isomics)
- Standalone FE Mesh Quality Viewer
- Mesh Quality Command Line Module
Mesh Improvement
Slicer3 Integration
Publications
- DeVries NA, Gassman EE, Kallemeyn NA, Shivanna KH, Magnotta VA, Grosland NM. Validation of phalanx bone three-dimensional surface segmentation from computed tomography images using laser scanning. Skeletal Radiol. 37(1):35-42, 2008.
- Gassman EE, Powell SM, Kallemeyn NA, Devries NA, Shivanna KH, Magnotta VA, Ramme AJ, Adams BD, Grosland NM. Automated bony region identification using artificial neural networks: reliability and validation measurements. Skeletal Radiol. 37(4):313-9, 2008.
- Grosland NM, Bafna R, Magnotta VA. Automated hexahedral meshing of anatomic structures using deformable registration. Comput Methods Biomech Biomed Engin. 2008.
- Magnotta V, Li W, Grosland N. Comparison of Displacement-Based and Force-Based Mapped Meshing. Workshop on Computational Biomechanics for Medicine at MICCAI 2008. Insight Journal, http://hdl.handle.net/1926/1490 , 2008.
Meetings and Events Specific to this Collaboration
- Vince Magnotta presented background information about the data acquisitioin and validation at the 2006 NA-MIC AHM (Slides: Validation of Bone Models using 3D Surface Scanning).
- November 7-8, 2006: Meshing Collaborator Project Visit to BWH
Resource Links
- Mesh warping project at the SPL - Steve Haker:
- Haker S, Warfield S, Tempany C. Landmark-Guided Surface Matching and Volumetric Warping for Improved Prostate Biopsy Targeting and Guidance. Proceedings of the 7th International Conference on Medical Image Computing and Computer-Assisted Intervention, MICCAI 2004, LNCS 3217, pp. 267-275, 2004.