Difference between revisions of "Measuring Alcohol Stress Interaction"
From NAMIC Wiki
| (5 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Projects List]] | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Projects List]] | ||
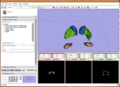
Image:MonkeyProject09Slicer.png|Segmentation of Vervet Subcortical Structures | Image:MonkeyProject09Slicer.png|Segmentation of Vervet Subcortical Structures | ||
| + | |||
</gallery> | </gallery> | ||
| Line 35: | Line 36: | ||
* [http://svn.na-mic.org/NAMICSandBox/trunk/MRIBiasCorrectionFilter/ MRIBiasCorrectionFilter] module by Nicolas Rannou available in NAMIC SandBox, initial testing on rhesus data done | * [http://svn.na-mic.org/NAMICSandBox/trunk/MRIBiasCorrectionFilter/ MRIBiasCorrectionFilter] module by Nicolas Rannou available in NAMIC SandBox, initial testing on rhesus data done | ||
* GWE set up on SPL Cluster for registration parameter exploration | * GWE set up on SPL Cluster for registration parameter exploration | ||
| + | * ChangerTracker used to detect change between hippocampus of State0 and State1 of first vervet subject. | ||
| + | {| | ||
| + | |[[Image:ChangeTracker_3dScreenShot_0.png|thumb|220px|ChangerTracker output for hippocampus changes between state0 and state1]] | ||
| + | |||
| + | |} | ||
| + | |||
| + | * MRIBiasCorrection Module parameter exploration being set up in GWE of SPL cluster. | ||
| + | {| | ||
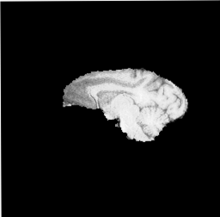
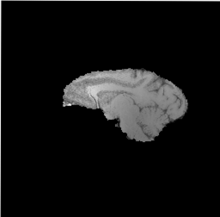
| + | |[[Image:Hugo_ICC_SagSlice128_BiasC_5.nii.base.png |thumb|220px|Before bias correction ]] | ||
| + | |[[Image:Hugo_ICC_SagSlice128_BiasC_5.nii.diff.png|thumb|220px|After bias correction]] | ||
| + | |} | ||
| + | <h3>In Progress</h3> | ||
| + | * Working with Xioadong to adapt SkullStripper Module for Vervet Images | ||
| + | * Explore use of ACPC align prior to rigid registration. | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 14:29, 26 June 2009
Home < Measuring Alcohol Stress Interaction
Key Investigators
- VT: Chris Wyatt, Vidya Rajagopalan
- BWH: Andrey Fedorov, Ron Kikinis, Nicolas Rannou, Sylvain Jaume
Objective
- To improve cortical segmentation of Vervet segmentation Results
- Use Change tracker module in Slicer to study hippocampal changes between subjects at two time points
Approach, Plan
- Improve rigid registration results by optimizing parameters
- Add bias correction to workflow to improve segmentation results
- Study hippocampal changes between subjects at two time points using Change Tracker
Progress
- ChangeTracker augmented with the capability to accept externally-defined segmentation (ChangeTrackerVT in NAMIC Sandbox)
- MRIBiasCorrectionFilter module by Nicolas Rannou available in NAMIC SandBox, initial testing on rhesus data done
- GWE set up on SPL Cluster for registration parameter exploration
- ChangerTracker used to detect change between hippocampus of State0 and State1 of first vervet subject.
- MRIBiasCorrection Module parameter exploration being set up in GWE of SPL cluster.
In Progress
- Working with Xioadong to adapt SkullStripper Module for Vervet Images
- Explore use of ACPC align prior to rigid registration.
References
- NA-MIC NCBC Collaboration: Measuring alcohol and stress interaction
- EM Segment project by Sylvain Jaume and Nicolas Rannou
- Tustison N., Gee J. "N4ITK: Nick's N3 ITK Implementation For MRI Bias Field Correction" InsightJournal, 2009 January - June, [1]