Difference between revisions of "CTSC Ellen Grant, CHB"
| Line 94: | Line 94: | ||
== Target Processing Workflow (Step 3) == | == Target Processing Workflow (Step 3) == | ||
| − | '''Step 3:''' | + | '''Step 3:''' |
* Execute query/download scripts | * Execute query/download scripts | ||
* Run processing locally, on cluster, etc. | * Run processing locally, on cluster, etc. | ||
Revision as of 03:10, 28 July 2009
Home < CTSC Ellen Grant, CHBBack to CTSC Imaging Informatics Initiative
Contents
Mission
Use-Case Goals
We will approach this use-case in three distinct steps, including Basic Data Management, Query Formulation and Processing Support.
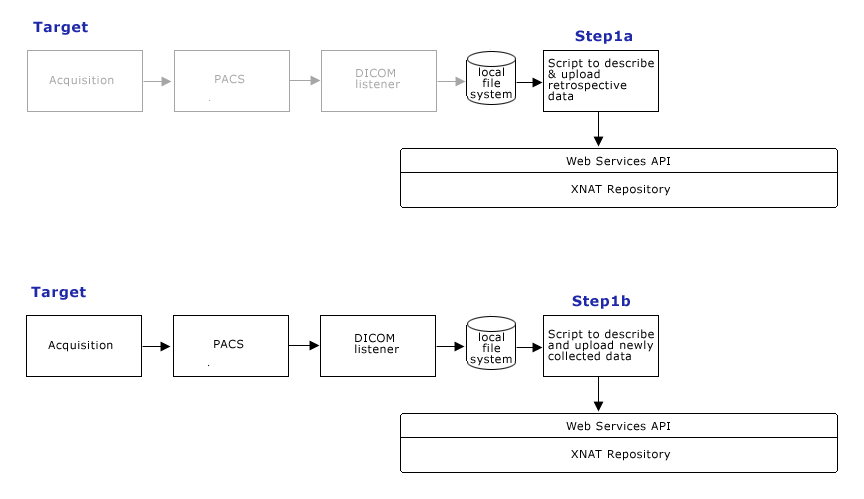
- Step 1: Data Management
- Step 1a.: Describe and upload retrospective datasets (roughly 1 terabyte) onto the CHB XNAT instance and confirm appropriate organization and naming scheme via web GUI.
- Step 1b.: Describe and upload new acquisitions as part of data management process.
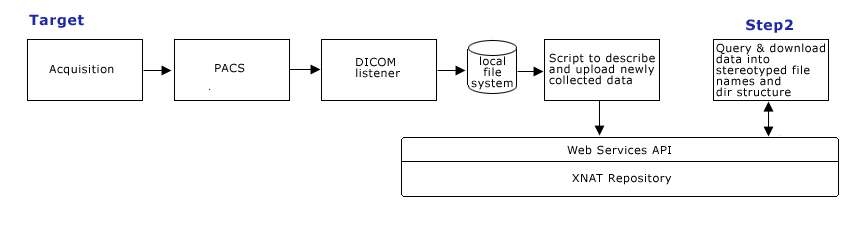
- Step 2: Query Formulation
- making specific queries using XNAT web services,
- data download conforming to specific naming convention and directory structure, using XNAT web services
- ensure all queries required to support processing workflow are working.
- Step 3: Data Processing
- Implement & execute the script-driven tractography workflow using web services,
- describe and upload results.
- ensure results are appropriately structured and named in repository, and queriable via web GUI and web services.
Participants
- sites involved: MGH NMR center, MGH Radiology, CHB Radiology
- number of users: ~10
- PI: Ellen Grant
- staff: Rudolph Pienaar
- clinicians
- IT staff
Outcome Metrics
Step 1: Data Management
- Visual confirmation (via web GUI) that all data is present, organized and named appropriately
- other?
Step 2: Query Formulation
- Successful tests that responses to XNAT queries for all MRIDs given a protocol name match results returned from currently-used search on the local filesystem.
- Query/Response should be efficient
Step 3: Data Processing
- Pipeline executes correctly
- Pipeline execution not substantially longer than when all data is housed locally
- other?
Overall
- Local disk space saved?
- Data management more efficient?
- Data management errors reduced?
- Barriers to sharing data lowered?
- Processing time reduced?
- User experience improved?
Fundamental Requirements
- System must be accessible 24/7
- System must be redundant (no data loss)
- Need a better client than current web GUI provides:
- faster
- PACS-like interface.
- image viewer should open in SAME window (not pop up a new)
- number of clicks to get to image view should be as few as possible.
Outstanding Questions
Plans for improving web GUI?
Data
Retrospective data consists of ~1787 studies, ~1TB total. Data consists of
- MR data, DICOM format
- Demographics from DICOM headers
- Subsequent processsing generates ".trk" files
- ascii text files ".txt"
- files that contain protocol information
Workflows
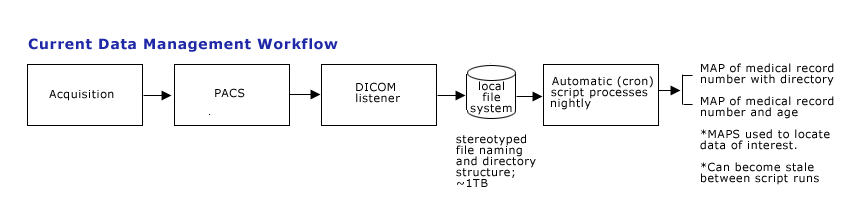
Current Data Management Process
DICOM raw images are produced at radiology PACS at MGH, and are manually pushed to the PACS hosted on KAOS resided at MGH NMR center. The images are processed by a set of PERL scripts to be renamed and re-organized into a file structure where all images for a study are saved into a directory named for the study. DICOM images are currently viewed with Osiris on Macs.
Target Data Management Process (Step 1)
Step 1: Develop an Image Management System for BDC (IMS4BDC) with which at least the following can be done:
- Move images from MGH (KAOS) to a BDC machine at Children's
- Step 1a: Import legacy data into IMS4BDC from existing file structure and CDs
- Step 1b: Write scripts to execute upload of newly acquired data.
Target Query Formulation (Step 2)
Step 2. Develop Query capabilities using scripted client calls to XNAT web services, such as:
Show all subjectIDs scanned with protocol_name = ProtocolName Show all diffusion studies where patients ages are < 6
- Scripting capabilities: Scripts need to query and download data into appropriate directory structure, and support appropriate naming scheme to be compatible with existing processing workflow.
Target Processing Workflow (Step 3)
Step 3:
- Execute query/download scripts
- Run processing locally, on cluster, etc.
- Describe & upload processing results
- Share images with clinical physicians
- Export post-processed data back to clinical PACS
Other Information
Rudolph has worked with XNAT support group at Harvard.
- 7/25/09 - Rudolph and Wendy are beginning experiments to upload representative data and metadata to CHB's XNAT instance.