Difference between revisions of "2010 WinterProject Week MRSIModule"
(Created page with '__NOTOC__ <gallery> Image:PW-SLC2010.png|Projects List Image:Mrsi parametric-map-NAA 3d.jpg|MRSI metabolite maps visualized in Slicer Image:…') |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | ||
| + | Image:Spectral fitting mrs.png|Fitting metabolite models to the MRS signal of tumorous brain tissue (top: baseline removal; bottom: resonance line model fits) | ||
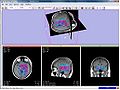
Image:Mrsi parametric-map-NAA 3d.jpg|MRSI metabolite maps visualized in Slicer | Image:Mrsi parametric-map-NAA 3d.jpg|MRSI metabolite maps visualized in Slicer | ||
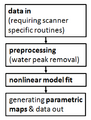
| − | Image: | + | Image:Workflow_MRSI_signal_processing.png| ''Results'': Workflow for the MRSI signal processing. Each processing step is to be realized as an individual external module. |
</gallery> | </gallery> | ||
| Line 12: | Line 13: | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| − | <div style="width: | + | <div style="width: 30%; float: left; padding-right: 2%;"> |
<h3>Objective</h3> | <h3>Objective</h3> | ||
Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive diagnostic method used to determine the relative abundance of specific metabolites at arbitrary locations in vivo. Certain diseases -- such as tumors in brain, breast and prostate -- can be can be associated with characteristic changes in the metabolic level. | Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive diagnostic method used to determine the relative abundance of specific metabolites at arbitrary locations in vivo. Certain diseases -- such as tumors in brain, breast and prostate -- can be can be associated with characteristic changes in the metabolic level. | ||
| Line 19: | Line 20: | ||
</div> | </div> | ||
| − | <div style="width: | + | <div style="width: 30%; float: left; padding-right: 2%;"> |
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
Spectral fitting routines have been implemented, using a HSVD filter for water peak removal and baseline estimation, and a constrained non-linear least squares optimization for the fit of the resonance line models. Current fitting routines require, however, several external software libraries not to be distributed, installed and used easily. | Spectral fitting routines have been implemented, using a HSVD filter for water peak removal and baseline estimation, and a constrained non-linear least squares optimization for the fit of the resonance line models. Current fitting routines require, however, several external software libraries not to be distributed, installed and used easily. | ||
| − | We want to | + | We want to find directions for either replacing these third-party libraries -- e.g. by using standard optimization routines from Python being available in Slicer -- or for making them easily available to the user of the module. |
</div> | </div> | ||
| − | <div style="width: | + | <div style="width: 35%; float: left;"> |
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | We focused on the second option and investigated directions for using third party libraries from within Slicer. | ||
| + | |||
| + | * We adapted the current MRSI processing routine to be separable into several sub-routines, each of which to be usable as external CLI module and depending on few (or single) external libraries only. | ||
| + | |||
| + | * We worked towards integrating an exemplary subroutine -- together with its external library -- as an external CLI module, potentially to be distributed as a Slicer Extension. | ||
| + | |||
| + | Separate subroutines can be integrated using a Slicer Wizard in accordance with the signal processing workflow (see ''Results'' figure above). | ||
| + | |||
</div> | </div> | ||
Latest revision as of 16:12, 8 January 2010
Home < 2010 WinterProject Week MRSIModuleKey Investigators
- MIT: Bjoern Menze, Polina Golland
Objective
Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive diagnostic method used to determine the relative abundance of specific metabolites at arbitrary locations in vivo. Certain diseases -- such as tumors in brain, breast and prostate -- can be can be associated with characteristic changes in the metabolic level.
The objective of the current project is to develop a module proving the means for the processing and visualization of MRSI -- and thus for a joint analysis of magnetic resonance spectroscopic images together with other imaging modalities -- in Slicer.
Approach, Plan
Spectral fitting routines have been implemented, using a HSVD filter for water peak removal and baseline estimation, and a constrained non-linear least squares optimization for the fit of the resonance line models. Current fitting routines require, however, several external software libraries not to be distributed, installed and used easily.
We want to find directions for either replacing these third-party libraries -- e.g. by using standard optimization routines from Python being available in Slicer -- or for making them easily available to the user of the module.
Progress
We focused on the second option and investigated directions for using third party libraries from within Slicer.
- We adapted the current MRSI processing routine to be separable into several sub-routines, each of which to be usable as external CLI module and depending on few (or single) external libraries only.
- We worked towards integrating an exemplary subroutine -- together with its external library -- as an external CLI module, potentially to be distributed as a Slicer Extension.
Separate subroutines can be integrated using a Slicer Wizard in accordance with the signal processing workflow (see Results figure above).