Difference between revisions of "Python Stochastic Tractography Tutorial"
| (10 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
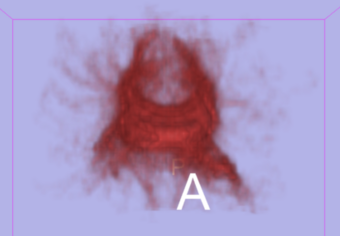
|[[Image:Cc_01.png|thumb|340px|Model of the Corpus Callosum from Stochastic Tractography (coronal) ]] | |[[Image:Cc_01.png|thumb|340px|Model of the Corpus Callosum from Stochastic Tractography (coronal) ]] | ||
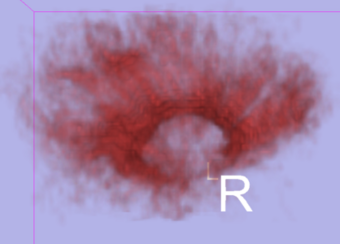
|[[Image:Cc_02.png|thumb|340px|Model of the Corpus Callosum from Stochastic Tractography (sagittal) ]] | |[[Image:Cc_02.png|thumb|340px|Model of the Corpus Callosum from Stochastic Tractography (sagittal) ]] | ||
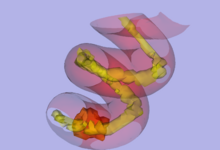
| − | |[[Image: | + | |[[Image:helix_withsmoothing.png|thumb|220px|Stochastic Tractography on Phantom]] |
|} | |} | ||
| Line 10: | Line 10: | ||
===Key Investigators=== | ===Key Investigators=== | ||
*PI: Marek Kubicki (kubicki at bwh.harvard.edu) | *PI: Marek Kubicki (kubicki at bwh.harvard.edu) | ||
| − | *DBP2 Investigators: Sylvain Bouix, Yogesh Rathi, Julien de Siebenthal, RA Doug Terry | + | *DBP2 Investigators: Sylvain Bouix, Yogesh Rathi, Julien de Siebenthal, RA Doug Terry, Andrew Rausch |
*NA-MIC Engineering Contact: Brad Davis, Kitware | *NA-MIC Engineering Contact: Brad Davis, Kitware | ||
*NA-MIC Algorithms Contact: Polina Gollard, MIT | *NA-MIC Algorithms Contact: Polina Gollard, MIT | ||
| + | *NAC: C-F Westin, BWH/Harvard Medical School | ||
===Tutorial=== | ===Tutorial=== | ||
| − | *[[Media: | + | *[[Media:IJData.tar.gz|Training Dataset]] |
| − | *[[Media: | + | *[[Media:Stochastic_June09_1.ppt|Training Presentation]] |
*[[Media:Helix.zip|Sample Helix Dataset]] | *[[Media:Helix.zip|Sample Helix Dataset]] | ||
Latest revision as of 18:12, 3 February 2010
Home < Python Stochastic Tractography Tutorial Return to Project Week Main Page |
Key Investigators
- PI: Marek Kubicki (kubicki at bwh.harvard.edu)
- DBP2 Investigators: Sylvain Bouix, Yogesh Rathi, Julien de Siebenthal, RA Doug Terry, Andrew Rausch
- NA-MIC Engineering Contact: Brad Davis, Kitware
- NA-MIC Algorithms Contact: Polina Gollard, MIT
- NAC: C-F Westin, BWH/Harvard Medical School
Tutorial
Objective
To address the uncertainty of deterministic tractography methods, stochastic tractography methods have been developed to quantify the uncertainty associated with estimated fibers (Bjornemo et al., 2002). Method uses a propagation model based on stochastics and regularization, which allows paths originating at one point to branch and return a probability distribution of possible paths. One can generate thousands of fibers starting in the same point by sequentially drawing random step directions. This gives a very rich model of the fiber distribution, as contrasted with single fibers produced by conventional tractography methods. Moreover, from a large number of sampled paths, probability maps can be generated, providing better estimates of connectivity between several anatomical locations.
Approach, Plan
This tutorial will walk users through the multiple stages of the python stochastic tractography module, where the inputs are a DWI and a region of interest labelmap. This will include:
- Loading the Volumes
- Extracting DWI and getting Baseline DWI threshold levels
- Using features of the module including smoothing, brain mask generation, and other settings.
- Generating a Connectivity Map, which is able to be thresholded and customized.
Progress
- A tutorial has been developed and data is available for tutorial.
- We have started the project of investigating Arcuate Fasciculus using Stochastic Tractography. This structure is especially important in both VCFS and schizophrenia, as it connects language related areas (Brocka and Wernicke's), and is involved in language processing quite disturbed in schizophrenia patients. It also can not be reliably traced using deterministic tractography.