Difference between revisions of "Projects:StructuralAndDWIPipeline"
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[Algorithm:Utah|Utah Algorithms]] | + | Back to [[Algorithm:Utah|Utah Algorithms]] |
__NOTOC__ | __NOTOC__ | ||
= A Framework for Joint Analysis of Structural and Diffusion MRIs = | = A Framework for Joint Analysis of Structural and Diffusion MRIs = | ||
| Line 24: | Line 24: | ||
* "Structural Image Preprocessing" | * "Structural Image Preprocessing" | ||
| − | + | Preprocess structural images to remove skull, correct bias field, normalize intensities, and segment tissue classes (to provide a white matter mask). | |
* "Group Atlas" | * "Group Atlas" | ||
| − | + | Build a structural atlas from all sub jects’ T1 images. Seed regions for tract endpoints are manually delineated in the structural atlas and then mapped | |
from the atlas to each individual. Automatically segment white matter tracts and quantify diffusion properties | from the atlas to each individual. Automatically segment white matter tracts and quantify diffusion properties | ||
using volumetric pathway analysis. | using volumetric pathway analysis. | ||
| Line 34: | Line 34: | ||
|[[Image:seeds.png|thumb|256px|The structural atlas built from the five T1 images with manually outlined | |[[Image:seeds.png|thumb|256px|The structural atlas built from the five T1 images with manually outlined | ||
frontal forceps seeds (left). The seeds mapped to each of the individual cases (right).]] | frontal forceps seeds (left). The seeds mapped to each of the individual cases (right).]] | ||
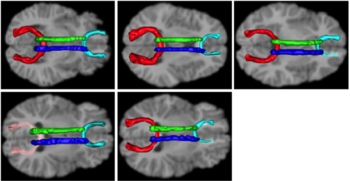
| − | |[[Image:tracts.png|thumb| | + | |[[Image:tracts.png|thumb|350px|Tracts on each of the individual cases.]] |
|} | |} | ||
| Line 41: | Line 41: | ||
* Utah: Ran Tao, Tom Fletcher, Ross Whitaker | * Utah: Ran Tao, Tom Fletcher, Ross Whitaker | ||
| + | |||
| + | = Publications = | ||
| + | |||
| + | * [http://www.na-mic.org/publications/pages/display?search=Projects%3AStructuralAndDWIPipeline&submit=Search&words=all&title=checked&keywords=checked&authors=checked&abstract=checked&sponsors=checked&searchbytag=checked| NA-MIC Publications Database on A Framework for Joint Analysis of Structural and Diffusion MRI] | ||
Latest revision as of 20:27, 11 May 2010
Home < Projects:StructuralAndDWIPipelineBack to Utah Algorithms
A Framework for Joint Analysis of Structural and Diffusion MRIs
Description
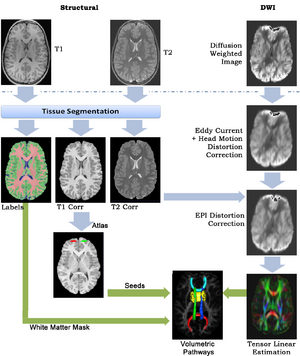
This framework addresses the simultaneous alignment and filtering of DWI images to correct eddy current artifacts and the subsequent alignment of those images to structural, T1 MRI to correct for susceptibility artifacts, and this paper demonstrates the importance of performing these corrections. It also shows how a T1-based, group specific atlas can be used to generate grey-matter regions of interest that can drive subsequent connectivity analyses. The result is a system that can be combined with a variety of tools for MRI analysis for tissue classification, morphometry, and cortical parcellation.
- Eddy Current Correction
We implemented the diffusion weighted image (DWI) registration model from the paper of G.K.Rohde et al.
Patient head motion and eddy currents distortion cause artifacts in maps of diffusion parameters computer from DWI. This model corrects these two distortions at the same time including brightness correction.
- "Structural Image Preprocessing"
Preprocess structural images to remove skull, correct bias field, normalize intensities, and segment tissue classes (to provide a white matter mask).
- "Group Atlas"
Build a structural atlas from all sub jects’ T1 images. Seed regions for tract endpoints are manually delineated in the structural atlas and then mapped from the atlas to each individual. Automatically segment white matter tracts and quantify diffusion properties using volumetric pathway analysis.
Key Investigators
- Utah: Ran Tao, Tom Fletcher, Ross Whitaker