Difference between revisions of "ITK GPAC level set"
Arnaudgelas (talk | contribs) |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * University of Missouri: K. Palaniappan | + | * University of Missouri: Ilker Ersoy, Filiz Bunyak, K. Palaniappan |
* Harvard Medical School: Kishore Mosaliganti, Sean Megason | * Harvard Medical School: Kishore Mosaliganti, Sean Megason | ||
| + | ==Project== | ||
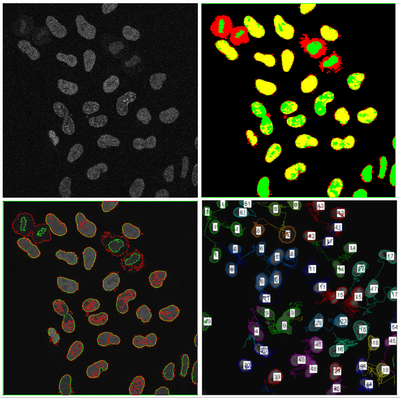
| + | [[File:Multiphase-GPAC-HeLa-segmentation.png|400px|thumb|left|4-phase cell segmentation]] | ||
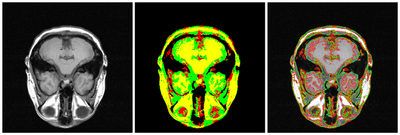
| + | [[File:MRI-multiphase-gpac.png|400px|thumb|left|4-phase MRI segmentation]] | ||
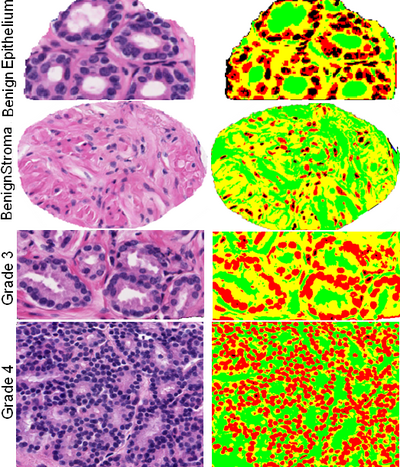
| + | [[File:Histopathology_mvls_4class.png|400px|thumb|left|Vector 4-phase histopathology segmentation]] | ||
| + | [[File:Histopathology_GVD2_grade4_2_zoom.png |400px|thumb|left|Nuclei detection using multiphase for Grade4 prostate carcinoma]] | ||
| + | |||
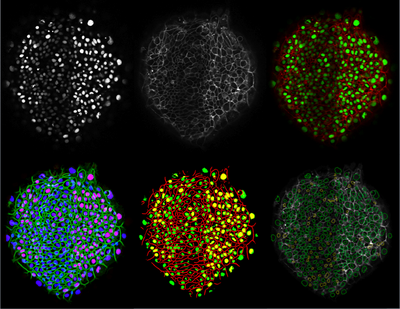
| + | [[File:Zebrafish-nuclei-membrane-channel-multiphase.png |400px|thumb|left|4-phase nuclei segmentation result using fused (weighted) nuclei plus membrane channels]] | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| − | <div style="width: | + | |
| + | <div style="width: 40%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| + | Multiphase level sets efficiently segment multiclass multiobject images. Current ITK level sets support the N-level set approach to handle N-objects. Multiphase methods can be applied to a variety of underlying level set energy functions including Chan-Vese, graph partitioning active contours (GPAC), 4-color level sets, and hybrid approaches. | ||
</div> | </div> | ||
| − | <div style="width: | + | <div style="width: 40%; float: left; padding-right: 3%;"> |
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| + | Extend the current ITK level set class to handle multiphase level sets with support for two, three or a general number of level sets cases. Multiphase methods model <math>N</math> classes using <math>\log N</math> phases. Each phase can be used to segment any number of objects. We plan to develop a mechanism for multiphase coupling of multichannel data (i.e. nuclear and membrane channels) with scalar or vector-based energy functions. We plan to include support for 2D and 3D multiphase curve and surface evolution. | ||
</div> | </div> | ||
| Line 17: | Line 27: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | Preliminary multiphase Chan-Vese level set segmentation algorithm has been developed using a Matlab reference implementation. Test cases with synthetic and microscopy imagery have been completed. Need to finish implementation for N-classes, extension to 3D and develop suite of test cases. | ||
| + | </div> | ||
| + | <h3>NA-MIC Progress</h3> | ||
| + | * Finished multiphase implementation and created preliminary test data for validation of results. | ||
| + | * Debugging of multiphase update equations for the general N-level set <math>2^N</math> phase case still continuing. | ||
| + | * Need to add test cases in ITK for specific 4-phase and 8-phase level set evolution. | ||
</div> | </div> | ||
| − | < | + | |
| + | ==Resources== | ||
| + | * Source code : | ||
| + | |||
| + | We shall use [http://git-scm.com/ git] for version control : | ||
| + | A small introduction to git : [http://sourceforge.net/apps/trac/gofigure2/wiki/GIT here] | ||
| + | * Data sets: | ||
| + | HeLa cell cycle phase analysis from Cristina Cardoso (Technical Univ of Darmstadt). | ||
| + | Prostate carcinoma from Michael Feldman (Univ of Pennsylvania). | ||
| + | Zebrafish embryogenesis 3D confocal multichannel imagery from Sean Megason. | ||
| + | <div style="width: 97%; float: left;"> | ||
| + | |||
| + | ==Delivery Mechanism== | ||
| + | |||
| + | This work will be delivered as an: | ||
| + | |||
| + | #ITK Module | ||
| + | #Slicer Module (possibly) | ||
| + | |||
| + | ==References== | ||
| + | *F. Bunyak, A. Hafiane, K. Palaniappan, Histopathology tissue segmentation by combining fuzzy clustering with multiphase vector level sets. Software Tools and Algorithms for Biological Systems, Springer, 2010. | ||
| + | |||
| + | *I. Ersoy, F. Bunyak, V. Chagin, M. C. Cardoso, K. Palaniappan, “Segmentation and classification of cell cycle phases in fluorescence imaging”, Lecture Notes in Computer Science (MICCAI), Vol. 5762 (Part II), pp. 617-624, 2009. | ||
| + | |||
| + | *F. Bunyak, K. Palaniappan, V. Chagin, M. C. Cardoso, "Cell Segmentation in time-lapse fluorescence microscopy with temporally varying sub-cellular fusion protein patterns", 31st Int. Conf. IEEE Engineering in Medicine and Biology Society (EMBC), Minneapolis, Minnesota, Sep. 2009, pp. 1424-1428. | ||
| + | |||
| + | *K. Palaniappan, F. Bunyak, S. Nath, J. Goffeney, “Parallel processing strategies for cell motility and shape analysis”, High-Throughput Image Reconstruction and Analysis, Ed. C.R. Rao and G. A. Cecchi, Artech, Chapter 3, 2009. | ||
| + | |||
| + | *F. Bunyak, K. Palaniappan, "Level set-based fast graph partitioning active contours using constant memory", Lecture Notes in Computer Science (ACIVS), Vol. 5807, pp. 145-155, 2009. | ||
| + | |||
| + | *A. Mosig, S. Jaeger, W. Chaofeng, I. Ersoy, S. K. Nath, K. Palaniappan, S.S. Chen, “Tracking cells in live cell imaging videos using topological alignments”, Algorithms in Molecular Biology, Vol. 4, 10p., 2009. | ||
| + | |||
| + | *F. Bunyak, K. Palaniappan, "Efficient segmentation using feature-based graph partitioning active contours", 12th IEEE Int. Conf. Computer Vision (ICCV), Sep. 29-Oct. 2 2009, Kyoto, Japan, pp. 873-880. | ||
| + | |||
| + | *S. K. Nath, K. Palaniappan, “Fast graph partitioning active contours for image segmentation using histograms”, EURASIP Journal on Image and Video Processing, 9p., 2009, Article ID 820986 (doi:10.1155/2009/820986). | ||
| + | |||
| + | *A. Hafiane, F. Bunyak, K. Palaniappan, “Clustering initiated multiphase active contours and robust separation of nuclei groups for tissue segmentation”, IEEE Int. Conf. Pattern Recognition, 2008. | ||
| + | |||
| + | *A. Hafiane, F. Bunyak, K. Palaniappan, “Fuzzy clustering and active contours for histopathology image segmentation and nuclei detection”, Lecture Notes in Computer Science (ACIVS), Vol. 5259, pp. 903-914, 2008. | ||
| + | |||
| + | *I. Ersoy, K. Palaniappan, “Multi-feature contour evolution for automatic live cell segmentation in time lapse imagery”, 30th IEEE Int. Conf. Engineering in Medicine and Biology (EMBC), pp. 371-374, 2008. | ||
<div style="width: 97%; float: left;"> | <div style="width: 97%; float: left;"> | ||
Latest revision as of 23:45, 24 June 2010
Home < ITK GPAC level setContents
Key Investigators
- University of Missouri: Ilker Ersoy, Filiz Bunyak, K. Palaniappan
- Harvard Medical School: Kishore Mosaliganti, Sean Megason
Project
Objective
Multiphase level sets efficiently segment multiclass multiobject images. Current ITK level sets support the N-level set approach to handle N-objects. Multiphase methods can be applied to a variety of underlying level set energy functions including Chan-Vese, graph partitioning active contours (GPAC), 4-color level sets, and hybrid approaches.
Approach, Plan
Extend the current ITK level set class to handle multiphase level sets with support for two, three or a general number of level sets cases. Multiphase methods model [math]N[/math] classes using [math]\log N[/math] phases. Each phase can be used to segment any number of objects. We plan to develop a mechanism for multiphase coupling of multichannel data (i.e. nuclear and membrane channels) with scalar or vector-based energy functions. We plan to include support for 2D and 3D multiphase curve and surface evolution.
Progress
Preliminary multiphase Chan-Vese level set segmentation algorithm has been developed using a Matlab reference implementation. Test cases with synthetic and microscopy imagery have been completed. Need to finish implementation for N-classes, extension to 3D and develop suite of test cases.
NA-MIC Progress
- Finished multiphase implementation and created preliminary test data for validation of results.
- Debugging of multiphase update equations for the general N-level set [math]2^N[/math] phase case still continuing.
- Need to add test cases in ITK for specific 4-phase and 8-phase level set evolution.
Resources
- Source code :
We shall use git for version control : A small introduction to git : here
- Data sets:
HeLa cell cycle phase analysis from Cristina Cardoso (Technical Univ of Darmstadt). Prostate carcinoma from Michael Feldman (Univ of Pennsylvania). Zebrafish embryogenesis 3D confocal multichannel imagery from Sean Megason.
Delivery Mechanism
This work will be delivered as an:
- ITK Module
- Slicer Module (possibly)
References
- F. Bunyak, A. Hafiane, K. Palaniappan, Histopathology tissue segmentation by combining fuzzy clustering with multiphase vector level sets. Software Tools and Algorithms for Biological Systems, Springer, 2010.
- I. Ersoy, F. Bunyak, V. Chagin, M. C. Cardoso, K. Palaniappan, “Segmentation and classification of cell cycle phases in fluorescence imaging”, Lecture Notes in Computer Science (MICCAI), Vol. 5762 (Part II), pp. 617-624, 2009.
- F. Bunyak, K. Palaniappan, V. Chagin, M. C. Cardoso, "Cell Segmentation in time-lapse fluorescence microscopy with temporally varying sub-cellular fusion protein patterns", 31st Int. Conf. IEEE Engineering in Medicine and Biology Society (EMBC), Minneapolis, Minnesota, Sep. 2009, pp. 1424-1428.
- K. Palaniappan, F. Bunyak, S. Nath, J. Goffeney, “Parallel processing strategies for cell motility and shape analysis”, High-Throughput Image Reconstruction and Analysis, Ed. C.R. Rao and G. A. Cecchi, Artech, Chapter 3, 2009.
- F. Bunyak, K. Palaniappan, "Level set-based fast graph partitioning active contours using constant memory", Lecture Notes in Computer Science (ACIVS), Vol. 5807, pp. 145-155, 2009.
- A. Mosig, S. Jaeger, W. Chaofeng, I. Ersoy, S. K. Nath, K. Palaniappan, S.S. Chen, “Tracking cells in live cell imaging videos using topological alignments”, Algorithms in Molecular Biology, Vol. 4, 10p., 2009.
- F. Bunyak, K. Palaniappan, "Efficient segmentation using feature-based graph partitioning active contours", 12th IEEE Int. Conf. Computer Vision (ICCV), Sep. 29-Oct. 2 2009, Kyoto, Japan, pp. 873-880.
- S. K. Nath, K. Palaniappan, “Fast graph partitioning active contours for image segmentation using histograms”, EURASIP Journal on Image and Video Processing, 9p., 2009, Article ID 820986 (doi:10.1155/2009/820986).
- A. Hafiane, F. Bunyak, K. Palaniappan, “Clustering initiated multiphase active contours and robust separation of nuclei groups for tissue segmentation”, IEEE Int. Conf. Pattern Recognition, 2008.
- A. Hafiane, F. Bunyak, K. Palaniappan, “Fuzzy clustering and active contours for histopathology image segmentation and nuclei detection”, Lecture Notes in Computer Science (ACIVS), Vol. 5259, pp. 903-914, 2008.
- I. Ersoy, K. Palaniappan, “Multi-feature contour evolution for automatic live cell segmentation in time lapse imagery”, 30th IEEE Int. Conf. Engineering in Medicine and Biology (EMBC), pp. 371-374, 2008.