Difference between revisions of "Seedings results comparison"
From NAMIC Wiki
Antonin07130 (talk | contribs) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
* Harvard Medical School: Antonin Perrot-Audet, Kishore Mosaliganti, Sean Megason | * Harvard Medical School: Antonin Perrot-Audet, Kishore Mosaliganti, Sean Megason | ||
| Line 29: | Line 24: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * Implemented | + | * Implemented algorithms in ITK: |
| − | ** | + | ** Radial voting, |
| − | ** | + | ** Multi-scale Distance Map weighted Laplacian of Gaussian. |
| + | * Created a collaboration framework using ITK: | ||
| + | ** Set of utilities for input images normalization | ||
| + | ** Developed a windowed local maxima filter | ||
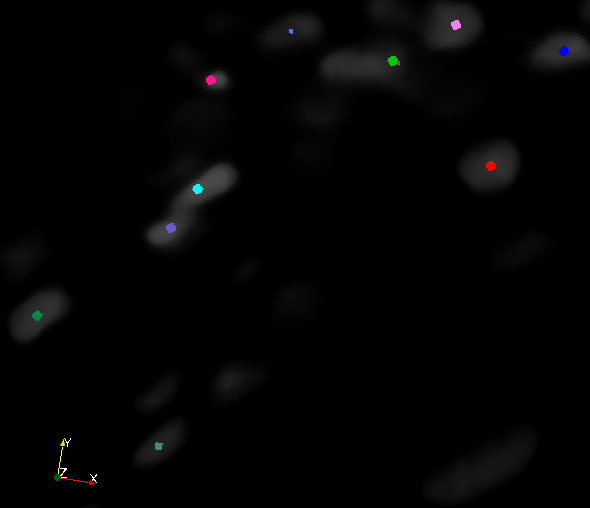
| + | * Evaluated and compared output of on synthetic 2D data & 3D Nuclei channel from Megason Lab | ||
** Multi-scale Distance Map weighted Laplacian of Gaussian. | ** Multi-scale Distance Map weighted Laplacian of Gaussian. | ||
| + | ** Radial voting | ||
| + | |||
| + | <h3>Next</h3> | ||
| + | * Implement state of the art seeding algorithms with ITK | ||
| + | ** gradient flow tracking | ||
| + | * Evaluate new algorithms | ||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
| + | ==Ressources== | ||
| + | * Source code : | ||
| + | git@github.com:antonin07130/NAMICSeeding.git | ||
| + | We shall use [http://git-scm.com/ git] for version control : | ||
| + | A small introduction to git : [http://sourceforge.net/apps/trac/gofigure2/wiki/GIT here] | ||
| + | * Data set : | ||
| + | Non public | ||
<div style="width: 97%; float: left;"> | <div style="width: 97%; float: left;"> | ||
Latest revision as of 14:06, 25 June 2010
Home < Seedings results comparisonContents
Key Investigators
- Harvard Medical School: Antonin Perrot-Audet, Kishore Mosaliganti, Sean Megason
- RPI: Badri Roysam, Raghav Padmanabhan
Project
Objective
- Improve segmentation algorithms initialization for nuclei detection in 3D fluorescent microscopy:
- get a better accuracy,
- improve computation speed.
Approach, Plan
- Compare different algorithms,
- Find a measure to evaluate different algorithm results,
- Fusion output of several algorithms.
Progress
- Implemented algorithms in ITK:
- Radial voting,
- Multi-scale Distance Map weighted Laplacian of Gaussian.
- Created a collaboration framework using ITK:
- Set of utilities for input images normalization
- Developed a windowed local maxima filter
- Evaluated and compared output of on synthetic 2D data & 3D Nuclei channel from Megason Lab
- Multi-scale Distance Map weighted Laplacian of Gaussian.
- Radial voting
Next
- Implement state of the art seeding algorithms with ITK
- gradient flow tracking
- Evaluate new algorithms
Ressources
- Source code :
git@github.com:antonin07130/NAMICSeeding.git
We shall use git for version control : A small introduction to git : here
- Data set :
Non public