Difference between revisions of "Projects:ARRA:SlicerEM"
Belhachemi (talk | contribs) |
Belhachemi (talk | contribs) |
||
| Line 31: | Line 31: | ||
***Created a Slicer extension for the Congealing project based on ( https://github.com/debonet/Congealing ) | ***Created a Slicer extension for the Congealing project based on ( https://github.com/debonet/Congealing ) | ||
***The extension is compiling successfully in debug mode on Linux and Mac | ***The extension is compiling successfully in debug mode on Linux and Mac | ||
| + | **EMSegmenter Updates: | ||
| + | *** Added new task 'Human Eye' | ||
| + | *** Separated EMSegmenter GUI (using KWWidgets) from EMSegmenter base code. | ||
| + | *** Future versions of the EMSegmenter GUI in Slicer4 (using Qt) can be linked against the same base code. | ||
| + | *** Fixed tests | ||
* 04/08/11 | * 04/08/11 | ||
**EMSegmenter Updates: | **EMSegmenter Updates: | ||
Revision as of 22:37, 15 April 2011
Home < Projects:ARRA:SlicerEMContents
Aim
The EMSegmenter is a state-of-the-art segmentation tool within 3D Slicer. User feedback has reported that clinicians are currently unable to tune the approach to their acquisition protocol, as the user interface is too complex. This proposal addresses this issue by redesigning the user interface, focusing on hiding the complexity of the underlying segmentation algorithm. If successful, this will enable clinicians to automatically segment their own medical scans, even if the corresponding acquisition protocol deviates from the default setting for which the EMSegmenter is optimized.
Research Plan
The EMSegmenter is the result of 15 years of research in medical image segmentation. This has lead to a user interface that exposes a rich set of parameters. These parameters allow the tuning of the EMSegmenter to a wide variety of acquisition sequences. However, tuning these parameters is quite challanging. In addition, Slicer currently does not provide any tools for generating atlases, which are a set of parameters characterizing each structure of interest. We propose to address these issues by creating two user-friendly modules: one for generating the atlases and one for tuning the EMSegmenter to a specific acquisition sequence.
The first module, called Atlas Generator, builds the atlases characterizing each structure of interest. The user simply specifies the training data which can be done via querying XNAT, a database targeted towards medical image analysis. The user also selects the type of information to be extracted from the data. Possible types are the shape, intensity, or relative position of the structures of interest across the training set. Based on this input, the tool automatically generates the atlas.
The second module, called EMSegmenter-Simple, consists of a simple work flow that enables users to adjust the EMSegmenter to their specific acquisition sequence. As part of this proposal, we will create a library of templates, which parametrizes the tool to segmentation tasks frequently ancountered by our user community. User simply adopt the tool to their acquisition scenario by first selecting the proper template. In the second step, the user modifies the value of important parameters of the template. We simplify the tuning of these parameters by providing an instant feedback mechanism. The feedback mechanism updates the automatic segmentation according to the change in the parameter setting. This will allow users to to get an intuitive understanding about impact of certain parameters on the algorithm. In addition, each entry field will be accompanied with a help text. We will also create detailed documentation about the user interface and publish a tutorial for each template.
The project is viewed as successful if a properly trained clinician is able to modify the templates to their acquisition sequence within an hour.
Events
- Advanced EMSegmenter Training : How to parametrize the tool
- Where: 1249 Bolyston Street, Boston, MA
- When: 10 am - 1 pm , Feb 23, 2010
Key Personnel
20% Kilian Pohl
95% Dominique Belhachemi
5% Daniel Haehn
Kitware (Project Manager: Jean-Christophe Fillion-Robin)
Documentation
- EMSegmenter documentation for Slicer 3.6
- Developer page
Progress
- 04/15/11
- Congealing Updates:
- Created a Slicer extension for the Congealing project based on ( https://github.com/debonet/Congealing )
- The extension is compiling successfully in debug mode on Linux and Mac
- EMSegmenter Updates:
- Added new task 'Human Eye'
- Separated EMSegmenter GUI (using KWWidgets) from EMSegmenter base code.
- Future versions of the EMSegmenter GUI in Slicer4 (using Qt) can be linked against the same base code.
- Fixed tests
- Congealing Updates:
- 04/08/11
- EMSegmenter Updates:
- Changed the default colormap to "GenericAnatomyColors.txt"
- Added new commandline option to switch on/off particular preprocessing steps (--TaskPreProcessingSetting="|C0|C0")
- Added commandline option (--atlasVolumeFileNames), so we can use the original mrml files, instead of duplicating them
- Prepared new CMTK extensions for all 3 platforms (Linux,Mac,Windows), compiled in Release mode with platform specific optimization flags. The speed up is 5x-6x
- Implemented a new task to call the AtlasCreator from within the EMSegmenter
- Atlas Creator Updates:
- detect labels automatically when using external invocation or the command line tool which was so far only possible in the GUI
- normalization of atlases does not force the float type anymore
- finished first version of CongealingCLI wrapper in terms of our un-biased groupwise registration effort and commited it to the Slicer3 trunk
- created documentation for the wrapper: http://www.slicer.org/slicerWiki/index.php/Modules:AtlasCreator:CongealingCLI
- added infrastructure to Slicer base to support the Invoke function in connection with KWWidgets
- handle events in MRML/GUI now properly and loop-safe
- only create new output folder if the configured one is not empty (now consistent between CLI and GUI)
- fixed a bug in connection with normalize when using the GUI
- fixed a possible crash when using the GUI and loading the generated atlases into Slicer
- added events to the comboboxes and write out MRML on change using the new Invoke method
- fixed possible memory leaks when using the GUI and exiting Slicer
- in pair fixed mode, also consider the defaultCase segmentation when combining to an atlas
- in pair fixed mode, also consider the defaultCase original when generating the normalized IntensityMap of all aligned images
- fixed possible error when selecting the defaultCase in the GUI: instead of the originals, a segmentation was selected
- added Testing, for now only a simple vtkMRMLAtlasCreatorNode test
- also, made sure to copy the TestData to the Slicer3 shared modules directory
- EMSegmenter Updates:
- 04/01/11
- Atlas Creator Updates:
- introduced vtkMRMLAtlasCreatorNode::InitializeByDefault() to set default values to the attributes of the MRML node
- re-structured the complete Graphical User Interface
- added events for all GUI elements to store the current state to MRML
- tested MRML import/load concept with AtlasCreator configurations successfully
- ran tests on SBIA cluster and identified need of mechanism to restart unsuccessful registration jobs
- optimized clean-up on Slicer shutdown
- added infrastructure to the 3D Slicer base to support listening to MRMLScene events without a corresponding calldata like the MRMLClose event
- PCA Analysis now fully integrated
- added new options to configure PCA modelling --pcaMaxEigenVectors X and --pcaCombine to the command line tool and the GUI
- Groupwise Registration effort:
- hands on Congealing sourcecode and successfully compiled it in different environments
- analyzed code and defined next steps with collaborators
- updated http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator#Priority_List
- Atlas Creator Updates:
- 03/25/11
- EMSegmenter Updates:
- The Atlas Creator can now be called from within a tcl script.
- Added a test script for this kind of invocation
- The results of the Atlas Creator are now written back to the MRML scene for further processing
- Performed static code analysis on the EMSegmenter
- Helped user (WUSTL) to find the optimal setting to segment their MRI data.
- Restructured the EMSegmenter tests
- Atlas Creator Updates:
- finished ITK implementation of combineToAtlas step (including normalization, re-casting..) called ACCombiner
- a lot of bug-fixing, tweaking and performance enhancing
- replaced the deprecated os.system calls to execute processes by the subprocess methods (new python API)
- re-structured advanced panel in GUI
- included PCA analysis capabilities using TCL (80%)
- available in the command line tool using --pca flag
- switched the CLI and the logic from the old configuration containers to use the vtkMRMLAtlasCreatorNode
- it is now possible to configure a MRML Node and invoke a launch event to start an AtlasCreator computation
- also, the API of vtkMRMLAtlasCreatorNode slightly changed to support the NormalizeTo and the DebugMode and DryrunMode settings
- moved ACCombiner to ACITKTools
- added new pure ITK tool in ACITKTools, called ACMeanImage to compute a meanImage of a bunch of images
- edited the AtlasCreator logic to use the ACMeanImage tool instead of computing it in the logic class
- integrated the ACMeanImage tool in the AtlasCreator cluster API
- as a result, finally all computation is moved from the headnode in cluster mode, even no images get loaded on the headnode
- added the feature to return as output as well the normalized intensity maps of aligned cases in dynamic and static mode
- coordinated effort on integrating Groupwise Registration (see http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator:Groupwise)
- EMSegmenter Updates:
- 03/18/11
- EMSegmenter Updates:
- ENH: Use Colormap="vtkMRMLColorTableNodeFileSlicer3_2010_Brain_Labels.txt"
- ENH: Update description of preprocessing checkboxes
- BUG: CMTK's registration/warp command cannot change the scalar type of the output volume, resample the atlas in an additional step
- ENH: Adding task for skull stripped brain images
- BUG: (BSplineToDeformationField) Send normal output to stdout instead of stderr , Use 'eval exec ' to execute a command
- BUG: Improve error handling for function calcDFVolumeNode
- Atlas Creator Updates:
- added fix to perform re-casting only when necessary to enhance performance
- included distributed computation of the last step: combineToAtlas including re-casting and normalization
- pure ITK code without XDISPLAY dependency
- as a result, all computation steps (registration, resampling, combining) now support a cluster environment
- solved problem of compiling CMTK on SPL cluster
- added the ability to normalize the atlases to 0..X instead of only 0..1
- supported in GUI and command line tool
- added fix to only register/resample images where segmentations exist and vice-versa
- created proof of concept to run the Atlas Creator from the EMSegmenter using MRML
- coordinated effort on integrating PCA Analysis (see http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator:PCA)
- coordinated effort on integrating Groupwise Registration (see http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator:Groupwise)
- EMSegmenter Updates:
- 03/11/11
- EMSegmenter Updates:
- Moved IO related functionality into base code
- ENH: Moved duplicated initialization code into a methed, bumped version number to 3.6.4-beta
- ENH: pass --fast --verbose options to each CMTK call
- ENH: Refactoring, move replicated code to new EMSegmentHelper file
- Adjusted Sandbox so it can use the current EMSegmenterHelper file
- Investigated a segfault during registration, CMTK's (nrrd reader) couldn't handle uint's , this is now fixed in the newest CMTK
- Atlas Creator Updates:
- Ran several tests and uploaded the data to http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator:Tests
- Identified and fixed problem with CMTK where certain labels were included in other atlases (see URL above, f.e. label 3 and label 9)
- Commited new version to the Slicer trunk including:
- Don't use Slicer temp dir anymore, use a temp dir in the output dir instead
- Refactored cluster mode to be based on a script
- Cluster script template (template.sh) is configurable for individual needs
- Added support for CMTK non-rigid registration
- De-activated multi-threading in cluster-mode for CMTK and BRAINSFit
- Different wait time for cluster mode for observing finished jobs
- Added date to print timestamps
- Distributed computation of Resampling
- Updated Priority List for new releases on http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator
- Started working on integrating PCA Analysis (see http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator:PCA)
- Started effort to include Groupwise Registration (see http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator:Groupwise)
- EMSegmenter Updates:
- 03/04/11
- Update MRI Human Brain Atlas (removed area around the brain)
- Tutorial-Simple updated for Slicer 3.6.3 release
- Tutorial-Advanced updated for Slicer 3.6.3 release
- Tutorial tested for 101 training page
- further work on the skull striper
- work on integrating I/O function into base
- Maintenance work for the Slicer 3.6.3 release
- Atlas Creator Updates:
- fixed a huge bug which modified the orientation of images during save/load cycle
- added feature to create a unique output directory automatically
- added guess background level functionality using EMSegment logic for CMTK and BRAINSFit
- released AtlasCreator version 0.21 as a fully usable module
- created test and example section for created atlases ( http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator:Tests )
- including fixed and dynamic registrations using CMTK and BRAINSFit on 50 cases
- planned coming releases v0.3 and v0.4 http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator#Priority_List
- installed Slicer Trunk on SPL cluster
- commited required fixes to Slicer base
- documented on SPL Intweb ( https://intweb.spl.harvard.edu/Dell_Linux_Cluster#How_to_compile_3D_Slicer_3_on_the_cluster.3F )
- 02/25/11
- EMSegmenter Updates:
- CMTK: Added support for multiple scalartypes
- CMTK: set the backgroundLevel to get rid of the segmentation errors outside the target volume
- updated the MRI Human Brain atlas
- work on the skull striper
- use qsub for running Slicer tools in the cluster (batch mode)
- use xvfb for Slicer tools which use the X protocol
- Atlas Creator Updates:
- introduced vtkMRMLAtlasCreatorNode
- It is possible to create a new AtlasCreator MRML Node and configure it
- the node can then be used to trigger an Atlas Creator computation by firing a Launch event
- introduced CMTK integration
- command line interface of the AtlasCreator supports adding the "--cmtk" flag to the command
- CMTK gets automatically detected, if installed as a Slicer extension
- optimized the handling of Double and Float images with BRAINSFit
- fixed and dynamic registration ran successfully on the SBIA cluster
- implemented the ability to listen to MRMLScene events in the Slicer3 Python Framework (Slicer Base)
- this enables listening to custom MRML Node events
- created small example on Slicer wiki: http://www.slicer.org/slicerWiki/index.php/Slicer3:Python:ProcessMRMLEvents
- provided mrml scene example for a fixed registration with 6 cases ( http://wiki.na-mic.org/Wiki/index.php/File:SceneFixed6Cases.zip )
- fixed bug in the Slicer3 Python Framework (Slicer Base) to exit Slicer correctly after using --evalpython
- created infrastructure to include C++ code in the AtlasCreator module using a GUI-less module approach
- included vtkImagePCAFilter and vtkImageKilianDistanceTransform as C++ filters successfully
- fixed overflow problem in connection with dynamic registration
- added the feature to remove all used temporary resources (cleanup)
- enabled the AtlasCreator C++ Code in the EMSegmentTclInterpreter (to test PCA filters)
- additional smaller fixes
- introduced vtkMRMLAtlasCreatorNode
- EMSegmenter Updates:
- 02/18/11
- EMSegmenter Updates:
- Our Mac OS X Server is submitting 2 fresh nightly builds of the Slicer3/branch-3-6 and the Slicer3/trunk to the dashboard. The names are MORRICONE and MINKOWSKI.
- Slicer3 (with python support) has been successfully installed on UPenn's SBIA cluster. OS: CentOS release 5.5 (Final)
- Adjusted parameter for CMTK so that it can be used in a fast mode (below one hour) and in a accurate mode (multiple hours).
- EMSegmenter maintenance
- Atlas Creator Updates:
- created a new command line interface (python atlascreator.py)
- including help and examples
- details available at the website http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerEM:AtlasCreator#The_Atlas_Creator_Command_Line_Interface
- fixed a blocker, where the resampled images where not used for Atlas generation
- found a BRAINSFit error in connection with dynamic registration which only appears on SPL machines
- investigated how to use CMTK as an alternative to BRAINSFit
- identified a problem with Slicer3 --evalpython
- commited a fix for a memory leak error when running the Atlas Creator
- Enabled code for PCA computation on local Slicer, still in the works, commited some fixes to specific SVN
- created and included UML diagrams on Wiki site
- created and included screenshots on Wiki site
- template now gets loaded into Slicer as well.
- temporary files are now saved in a subdirectory with a unique name so the Atlas Creator can be run multiple files without moving anything.
- in cluster mode, multi threading is now disabled.
- created a new command line interface (python atlascreator.py)
- EMSegmenter Updates:
- 02/11/11
- Installed MacOS and WindowsXP test machines
- It is now possible to use a temporary directory with white spaces (this was a bug on Windows XP)
- Added manual sampling to EMSegmenter module
- Atlas Creator Updates:
- Generalized API to be able to run the Atlas Creator from external modules or using the Slicer launcher via command-line
- Used the opportunity to extensively comment the code
- Implemented Support for Parallelized Computation on a Cluster
- Finished Dynamic Registration against Mean of images
- Added mode to use existing transforms (f.e. to re-generate an Atlas)
- Added in-module help
- Smaller Fixes and Changes
- Merged latest version to Slicer 3.6.3 branch - ready for release
- Updated Projects:ARRA:SlicerEM:AtlasCreator
- Generalized API to be able to run the Atlas Creator from external modules or using the Slicer launcher via command-line
- 02/04/11
- option intermediate results directory is created for the user
- clarified the usage, helped collaborators with the EMSegmenter
- Windows specific runtime bug fixed, happened whenever we used an iterator two times.
- Included detailed verbose printout in EMSegmentCommandLine
- Added test results to MIDAS server
- Use CMake to compare segmentation results with precomputed segmentation results
- Further cleanup code (Number of registration flags simplified)
- Simplified MRML structure(removed nodes)
- Check if atlas files are properly downloaded to avoid FetchMI checksum checks
- Fixed bug in test EMSegCL_IntermediateResults: "UpdateScene: error getting 0th storage node"
- Updated template creator to new MRML structure (new MRML structure)
- Fixed the updated functionality for EMSegmenter
- also transferred fix to Extension wizard (was very quick)
- added new repository for 3.6.3 for EMSegment tasks
- fix for the new webserver index files and therefore do not need the index.html
- committed all code to 3.6.3 Branch and Trunk
- tested on Mac and Linux
- Atlas Creator Updates:
- Created diagram of Atlas Creator workflow
- Support for Normalization of output Atlases done
- Support for Selecting the Output Cast done
- Re-Designed API
- Focus on Cluster proposal
- Focus on external invocation (command line or 3rd party module in 3D Slicer)
- Introduced Configuration Concept
- Documentation, Diagram and Priority List updated at Projects:ARRA:SlicerEM:AtlasCreator
- 01/28/11
- Fixed a bug in the test 'intermediateResultsDirectory'. The test doesn't use the results file which is specified in the mrml scene. Instead it is using a test specific results file(specified in the CMake template).
- Fixed a Slicer4 bug : "ENH: Call the GeneralGauss(...) functions instead of the FastGauss(...) functions - this avoids type punning" Modules/EMSegment/Graph/vtkImageCurveRegion.cxx [15896]
- Fixed a Slicer4 bug : "BUG: Use a normal gauss function to avoid type-punning" QTModules/EMSegment/Widgets/vtkPlotGaussian.cxx
- Worked on the CT-Hand-Bone task
- Worked on a EMSegmenter specific skull stripper
- Atlas Creator Updates:
- Research and Proposal of running the Atlas Creator in a cluster environment
- Several GUI Fixes
- Support Dynamic Registration (in the works)
- Support for using existing Registrations by loading deformation fields (in the works)
- Support for Normalization of output Atlases (in the works)
- Support for Selecting the Output Cast (in the works)
- Merged Export of Deformation Fields and Export of Transforms
- 01/21/11
- Worked on new CT-Hand-Bone task
- Maintaining work on existing tasks
- Added new features to the Atlas Creator
- New wiki page Projects:ARRA:SlicerEM:AtlasCreator
- 01/14/11
- Created a test for the MRI-Human-Brain-Full-Parcellation task
- Worked on failing tests if compiling Slicer3 with Superbuild
- Worked on new CT-Hand-Bone task
- Worked with Wendy on new ideas how to make FetchMI more rubust
- 01/07/11
- Created new Atlas Creator module for Slicer3
- Created new Wiki page for Call for Datasets for the EM Segmenter Use Case Library
- 12/31/10
- Help debugging FetchMI bugs 1027 983
- Updated MRI-Human-Brain-Parcellation
- 12/24/10
- Add postprocessing to segmentation, which includes island removal and subparcellation of tissue classes
- 12/17/10
- Added down-sampled image data for testing purposes
- Added two tests for each task
- Added UPenn logo to the EMSegmenter acknowledgment section
- Work on FetchMI bugs
- Updated Wiki page for the task MRI-Human-Brain-Parcellation
- 12/10/10
- Added wrapper for SkullStripperCLI to GenericTask.tcl
- Updated CMTK extension, using now Torsten Rohlfing's repository: https://nitrc.org/svn/cmtk/tags/Slicer3
- Updated Advanced Tutorial based on Ron's comments
- Added functionality to threshold target image(s), negative values will be set to 0 so that the EMSegmenter can be used on those images
- 12/03/10
- Create a new Wiki page for the EMSegmenter Task Library [1]
- Updated task files and documentation
- MRI-Human-Brain
- MRI-Human-Brain-Parcellation
- Non-Human-Primate
- Fixed a apache server issue to update tasks in Slicer 3.6.2
- 11/26/10
- Fixed bug: when creating new task that registration was performed regardless if it was selected or not
- Worked on test cases for 'Non Human Primate' pipeline
- 11/19/10
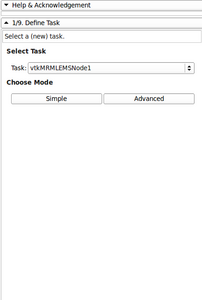
- Created a new EMSegmenter Tutorial (simple mode) - (for quick results, no need to change parameters, this will be useful to clinicians)
- Created a new EMSegmenter Tutorial (advanced mode) - for experienced user of the EMSegmenter
- Created the new 'MRI Human Brain Parcellation' pipeline and added to 3D Slicer 3.6.2 (collaborators: Padmapriya Srinivasan and Sylvain Bouix, PNL BWH)
- 11/12/10
- Restructuring EMSegmenter's Tasks folder to support multiple pipelines
- Start working on a 'MRI Human Brain Parcellation' pipeline (collaborators: Padmapriya Srinivasan and Sylvain Bouix, PNL BWH)
- Start working on a 'Non Human Primate' pipeline (collaborators: Andriy Fedorov, BWH)
- Slicer 3.6.2-RC2 maintenance
- 11/05/10
- software testing for Slicer 3.6.2 release
- Added new test for option --intermediateResultsDirectory
- Fixed bug 1023: MRML import doesn't read copied mrml scenes reliable
- Fixed bug 1025: Slicer crashes when BRAINSFit returns error
- 10/29/10
- Pop up error message when performing segmentation
- Add CMTK in Generic.tcl
- BUG: use .mat extension for transformation files" GenericTask.tcl
- Bug fixing with Slicer Base Code developers
- 10/23/10
- Bug fixes for Slicer 3.6.2 release
- Memory leak fixed
- comment-out unused code in MRI-Human-Brain.tcl
- 10/15/10
- Added function for automatically generating Template File
- Added function for downloading task files from web
- Added CMTK to extension module
- One can assign now tcl file when creating a new task
- Added class name to all tabs
- Tested command line
- Created virtual machines to submit tests reports to the dashboard
- Created Popup window when an error message appears in pre-processing and segmentation
- EMSegmenter works now with different image data types (short, float,...)
- Removed several bugs in mrml structure
- 10/8/10
- Added the --tmp_dir option to the start up allowing one to specify slicers temporary directory
- Fixed the leaks in the nightly tests
- 10/1/10
- Removed command line nodes from mrml scene after the command line is executed
- Added functionality to call N4ITKBiasFieldCorrectionCLI from EMSegmentCommandLine
- Added label - in addition to the tree - to show the selected anatomical tree node
- Patched KWWidgets to specify configuration directory (like /tmp/.3D\ Slicer\ Version\ 3.6.2-betarc)
- Working on parallel execution of multiple instances of the EMSegmentCommandLine
- 9/24/10
- Added check to make sure that the user cannot delete the root node
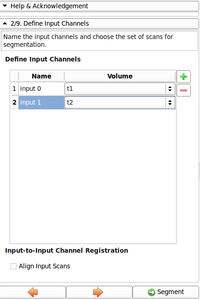
- Added a warning to avoid ambiguous input channel names
- Added check for emtpy 'Task Name'
- Added a label to show the selected node during specifying the priors
- Alpha value was not correctly transfered from node to vtkImageEMLocalSegmenter
- 9/17/10
- 969 fixed: missing quick timeout if connection to web resource is blocked (closed, see commit 14981+14982)
- 954 fixed:EMSegmentCommandLine throws errors if started from a current working directory different than ./Slicer3-build
- 948+958 fixed:EMSegmentCommandLine segfaults in cleaning up mrmlScene step
- 959 fixed: EMSegmenter ignores option --disableMultithreading
- Added progress dialogs for pre-processing and segmentation
- Test GUI on Linux, Windows, and Apple for Slicer 3.6.2 release
- 9/10/10
- Investigating a critical segfault bug 948+958
- Reviewing use of the extension manager for our needs
- Resolved bugs 970, 959, 944, 943
- 9/3/10
- Updated Image gallery
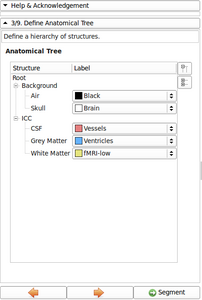
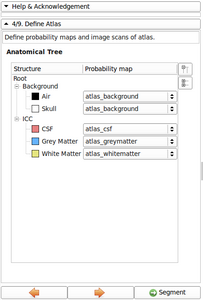
- Refine Anatomical tree widget (Probability map integrated, Icon associated with structure name, ...)
- Now possible to design step directly from QtDesigner
- Discussed integration of PNL kMeans pipeline into Slicer 3.6
- Discussed integration of CNL workflow as a EMSegmenter pipeline into Slicer 3.6
- Discussed differences between the SBIA cluster and the BWH cluster,
- Conclusion: SGE seems to be easier to handle than Lava
- Working on bug 959: EMSegmenter ignores option --disableMultithreading
- 8/27/10
- Integrated Workflow manager with QtModule
- Review Workflow manager API
- Fix bug related to PythonQt wrapping
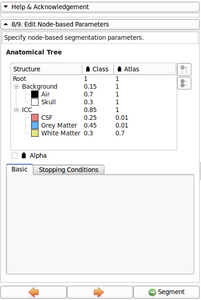
- Now possible to update atlas class weight, atlas weight and alpha value directly from the Anatomical tree
- Updated EMSegmenter related Wiki pages
- Testing EMSegmenter command line executable in a Sun Grid Engine (SGE) environment (branch 3.6 + trunk)
- working on related bugs 954, 955, 948
- changing some stderr output to stdout
- Updating EMSegmenter tutorial for Slicer 3.6
- 8/20/10
- Ported anatomical tree widget and list of input channel - The anatomical tree is generic enough and re-used.
- Added Image gallery
- 8/13/10
- Meet with Kilian - review panels and widgets and talked about which features should be improved, changed or omitted
- 8/6/10
- Now possible to enable wrapping of QtModule (Gui, UItools) in Slicer through CTK/PythonQt
- Discuss how TCL script interacted with the KWWidgets workflow manager
- 7/30/10
- The port of the Workflow manager is initiated
- Created Qt static panel skeleton corresponding to the step of MRI human brain workflow
- Ability to script task in python is something important. Re-prioritize the task.
- Discussed how dynamic panel could be ported.
- 7/23/10
- Reviewed milestone and list of widgets to port
- Created initial directory structure
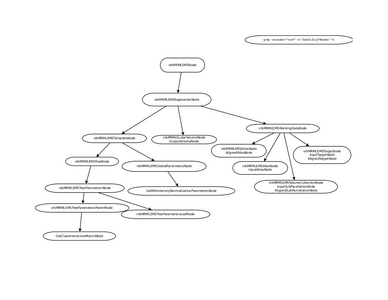
- Initiated the conversion of Graph/Algorithm/MRML code to separate shared module
- The port of existing TCL function related to processing is low priority
- 7/16/10
- First weekly meeting between Kitware and UPenn. Meeting will always be held after the Annotation/QT tcon.
- 7/9/10
- Remove bugs
- Set up contract with Kitware to port GUI to QT
- 7/2/10
- Hired Dominique Belhachemi
- Transfered grant from IBM to UPenn
- Wrote Progress Report
- 6/25/10
- Addressed all EMSegmenter bug in Mentis
- 6/18/10
- Revising task for MRI brain segmentation
- Started search for new hire
- Make EMSegmenter work with Slicer Superbuild
- 6/11/10
- Updated EMSegmenter documentation to reflect changes in Slicer 3.6 interface
- Fixed major bugs so that Slicer 3 does not crash when creating a new task
- 6/04/10
- Imbedded BRAINSFit registration in the pipeline
- 5/28/10
- Changed Simple version of EMSegmenter to work with new preprocessing structure
- 5/21/10
- Fixed class overview window
- 5/14/10
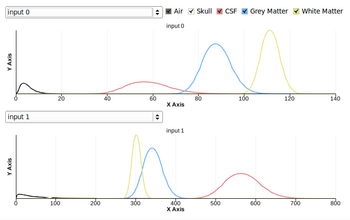
- Fixed Intensity graph distribution display
- 5/07/10
- Fixed EMSegmenter command line module
- 4/30/10
- Debugged EMSegmenter for Slicer 3.6 release
- 4/23/10
- Added segmentation button at each Step of the Wizard
- Integration of 3D/2D Bounding Box selection
- 4/16/10
- Updated Slicer 3.6 Documentation
- Modified workflow of Slicer 3.6
- 4/9/10
- Created simple interface for preprocessing
- 4/2/10
- Incorporated Bug fixes from Slicer 3.4 into trunk
- Removed bugs so that selecting tasks from a list properly works
- Rolled EMSegmenter back to Slicer version 3.4
- EMSegmenter Research environment now compiles under windows
- 3/26/10 Help users of EMSegment to adopt the tools to their needs
- 3/19/10 Manuscript was accepted by CVPR 2010 for poster presentation
- 3/12/10 Start organizing tutorials for EMSegmenter
- 3/05/10 Extended first step in EMSegmenter to include default task list
- 2/26/10 Removed bug so that api interface returns the same results as GUI version in Slicer2.6 . Trained members of CNL, BWH in segmenting lesions from MR brain images. Introduced CNL team to post processing tools for lesion segmentation.
- 2/19/10 Working with Andrey Fedorov begin_of_the_skype_highlighting end_of_the_skype_highlighting begin_of_the_skype_highlighting end_of_the_skype_highlighting begin_of_the_skype_highlighting end_of_the_skype_highlighting begin_of_the_skype_highlighting end_of_the_skype_highlighting begin_of_the_skype_highlighting end_of_the_skype_highlighting, BWH, to customize the EMSegmenter to non-human primate brain images
- 2/12/10 Presented Slicer to IBM Health Care Division
- 2/05/10 Transferring Responsibilities for the EMSegmenter in Slicer 3.5 from MIT to IBM
- 1/29/10 Migrated EMSegmenter Research Environment from Slicer2 to Slicer3
- 1/22/10 Created slides for NAC Site visit
- 1/15/10 Started to resolve bugs that requires collaboration between Kitware, BWH, and IBM team
- 1/8/10 Trained members from BWH and Virginia Tech on using EMSegmenter on Non-Human primate data
- 1/1/10 Addressed all major bugs listed in Mantis for Slicer Version 3.4
- 12/25/09 Setup Slicer environment for new hire
- 12/18/09 Hired Yong Zhang to help with maintenance of EMSegmenter
- 12/11/09 Learned about programming user interfaces in Qt
- 12/04/09 Interviewed applicant for Post Doc position to work on this project
- 11/29/09 Removed bugs related to updated CMake version
- 11/20/09 Organize bugs related to EMSegmenter
- 11/13/09 Tcon featuring the EMSegmenter - participants Andriy Fedorov, Sylvain Jaume, Stuart Wallace, Kilian Pohl - Result of discussion
- Sylvain will fix EMSEgmenter bugs Slicer 3.5
- Kilian will fix any EMSegmenter bugs in Slicer 3.4 and earlier
- Andriy is developing a segmentation pipeline for the Wake Forest Data
- Stuart will do testing of the EMSegmenter module
- 11/06/09 Meet with Jean-Christophe Fillion-Robin from Kitware to discuss integration of Qt in 3D Slicer
- 10/30/09 Organized Monthly TCon between EMSegmenter developers
- 10/23/09 Organized onsite interview , got in contact with Steve Pieper to discuss next steps, installed Slicer3
- 10/17/09 Started interviewing postdoc as well as solving several HR issues for hiring personal
Image gallery
- EM Segmenter - Port to Qt
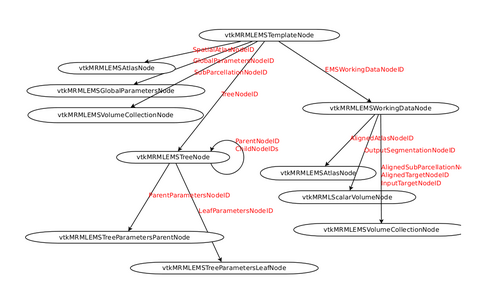
MRML Structure
- EM Segmenter - MRML Structure