Difference between revisions of "2012 Progress Report Science Wiki Version Engineering"
| Line 45: | Line 45: | ||
<em>Dan Marcus</em> | <em>Dan Marcus</em> | ||
| − | Efforts in the Data Management Platform have focused on developing an ergonomic user interface and internal networking logical to efficiently exchange data between Slicer and XNAT. | + | Efforts in the Data Management Platform have focused on (1) developing an ergonomic user interface and internal networking logical to efficiently exchange data between Slicer and XNAT and (2) expanding Slicer's support for importing from and exporting to local DICOM Objects and networked DICOM PACS. |
| + | |||
=== Major Developments === | === Major Developments === | ||
* ''User Interface:'' The XNAT development team is working with a web usability firm (Integrity St. Louis) to design and implement a web interface that can be deployed in Slicer 4's Qt framework for exploring data hosted in a remote XNAT data repository. Prototypes have been implemented and are currently being reviewed by stakeholders with the goal of a functioning interface by June, 2012. | * ''User Interface:'' The XNAT development team is working with a web usability firm (Integrity St. Louis) to design and implement a web interface that can be deployed in Slicer 4's Qt framework for exploring data hosted in a remote XNAT data repository. Prototypes have been implemented and are currently being reviewed by stakeholders with the goal of a functioning interface by June, 2012. | ||
* ''Networking logic:'' An alpha implementation of the infrastructure for actually exchanging data between XNAT and Slicer has been developed and revealed that significant additional work is required to handle parsing of MRML files within Slicer in the context of remotely hosted data. This work is now underway and will require several months to complete. | * ''Networking logic:'' An alpha implementation of the infrastructure for actually exchanging data between XNAT and Slicer has been developed and revealed that significant additional work is required to handle parsing of MRML files within Slicer in the context of remotely hosted data. This work is now underway and will require several months to complete. | ||
| − | * ''Use cases:'' Several use cases have been identified, including projects within the Quantitative Imaging Network (QIN), to drive the development of the Slicer/XNAT integration. | + | * ''Use cases:'' Several use cases have been identified, including projects within the Quantitative Imaging Network (QIN), to drive the development of the Slicer/XNAT integration. |
| + | * '''DICOM-RT and QIN support:''' Via DCMTK and custom classes, Slicer is being extended to support RT Plans, Images, Annotations, etc. Much of the support is being driving by the head-and-neck cancer core. Additional developments are being driven by an ongoing effort to integrate Slicer as an annotation module in the Quantitative Imaging Network (QIN). | ||
| + | * '''DICOM database and networking:''' Slicer DICOM support is approaching clinical quality in terms of speed of searching and IO by maintaining a database that indexes previously loaded DICOM objects. Via this database, it is no longer necessary to parse each object in order to search and/or load an entire series, study, or patient into Slicer. | ||
=== Plans === | === Plans === | ||
| − | The bulk of the effort in the coming year will continue to focus on the user interface and networking logic for remote data management. | + | The bulk of the effort in the coming year will continue to focus on (1) the user interface and networking logic for remote data management in XNAT and (2) import and export of Slicer results (i.e., entire MRML scenes) as DICOM objects. Regarding XNAT, as the initial development completes, we will turn to developing more efficient mechanisms for caching data locally and synchronizing local caches with remote repositories. Regarding importing and exporting Slicer data as DICOM objects, we are pursuing the concept of a '''DICOM Lollipop.''' In a DICOM Lollipop, a complete Slicer (MRML) Scene, with annotations, segmentations, viewing conditions, etc, can be saved as a binary payload in a standard DICOM object. In this manner, Slicer's data can be pushed/pulled from PACS for integration with and sharing across hospital workflows. |
== 5.2.4. Community Software Process == | == 5.2.4. Community Software Process == | ||
Revision as of 21:42, 20 April 2012
Home < 2012 Progress Report Science Wiki Version EngineeringContents
5.2. Engineering
Key Investigators
- Will Schroeder, Kitware
- Stephen R. Aylward, Kitware
- Steve Pieper, Isomics
- Jim Miller, GE Research
The Engineering component of the Computer Science Core (Core 1b) has been focusing on the infrastructure needed for the Algorithms component to implement their methods, and has been working closely with them so that the functionality we provide can serve to inspire new methods as well. Herein we provide more details regarding our accomplishments in those directions
5.2.1. End-user Platform: 3D Slicer
Steve Pieper
Release of 3D Slicer version 4.0 and 4.1
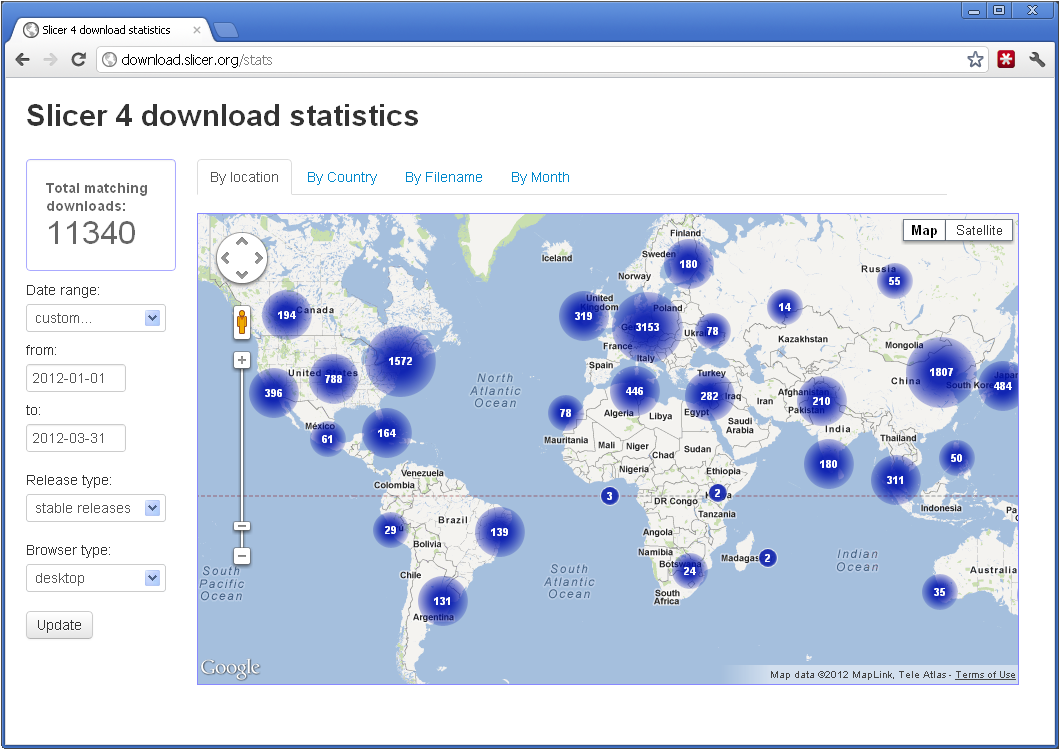
As shown in Figure 1, 3D Slicer version 4.0 (commonly called Slicer4) is having an immediate world-wide impact. Version 4.0, released at the Radiology Society of North America (RSNA) meeting in late November 2011, is the result of a major effort by the NA-MIC engineering cores, in collaboration with the wider Slicer community, to re-implement and streamline the software in response to feedback from NA-MIC DBPs and algorithm developers. A new 3D Slicer version 4.1 release, being finalized at the time of this writing, adds back many of the advanced features of Slicer3, notably the Extension system by which new functionality can be downloaded and installed independent of the main executable. As described more fully below, these sweeping changes required touching not only most of the code in Slicer, but also feeding important feature and bug fix changes back into the rest of the NA-MIC Kit and upstream libraries. As a result of these efforts, 3D Slicer in now an improved reference implementation of a modern medical image computing package and a strong foundation for research.
Figure 1: 3D Slicer version 4.0 end-user downloads during the first 3 months of 2012, a rate of 45,360 per year (over 100 downloads per day). Interactive geolocated download statistics are available at http://download.slicer.org/stats.
Major Developments and New Functionality in Slicer4
- Modern Cross-Platform Design Patterns: As a by-product of the port of the user interface from KWWidgets to Qt, a comprehensive suite of non-GUI OS abstractions and utility functions from Qt became available to support core application functions such as preference settings, multi-processing, application resource data. This was a direct benefit from the GUI port supported by an ARRA supplement to the Neuroimage Analysis Center, a NA-MIC collaborating P41 grant. Slicer4 supports native windows 64 bit environments and can be bundled into a standard Mac OS X application bundle.
- Efficiency and Robustness: Careful review of core data management and processing pipeline steps allowed us to remove much redundant processing. The result is that Slicer is easier for developers to understand and debug, and end users experience faster startup and more responsive behavior.
- DICOM Networking: Slicer4 includes a DICOM listener and DICOM Query/Retrieve capabilities for integration with standard clinical image management environments and workflows. For example, intraprocedural imaging obtained during image guided procedures can now be auto-routed to Slicer for analysis and navigation.
- Additional Features: Slicer4 includes: an improved flexible view layout system; a revised implementation of the Expectation Maximization (EM) Segmenter; faster hardware accelerated volume rendering; improved markups and annotations; improved atlas and model hierarchy support; a streamlined and revised diffusion MRI implementation.
Plans
With the introduction of the Slicer4 Extension system we plan to stabilize the core slicer distribution and move to less frequent releases with more of the algorithm innovation becoming available through as Extensions. This will allow further stabilization and streamlining of the core while speeding up the delivery of new technologies to end users for testing.
5.2.2. Computational Platform
Jim Miller
Efforts in the Computational Platform have focused on developing a general and flexible computing architecture and analysis platform to meet the needs of NA-MIC scientists and engineers as well as the larger medical imaging community.
Major Developments
- Interactive methods: The Editor module has supported interactive segmentation techniques that were deeply integrated with the Editor codebase. We have broadened the Slicer Extension mechanisms to support Editor Extensions, allowing interactive segmentation techniques to be developed separately from the Slice codebase. We have also refined the interaction patterns for interactive segmentation within the Editor and are starting to develop interactive registration techniques.
- Multivolume analysis: The infrastructure for Diffusion Weighted MRI (DWI) IO and visualization has been generalized to be used for other time varying acquisitions like Dynamic Contrast Enhanced MRI (DCE) and Gated Cardiac CT. Massively univariate processing of DCE to produce parametric maps is being developed as a Slicer Extension.
- Distributed computing: Slicer Execution Model modules (also known as Command Line Modules) are now available as Nipype tools, enabling local and distributed scripted execution of processing pipelines.
- Exploratory image analysis: Infrastructure for the interactive exploration of images and the relationships of features calculated over regions of images was developed, including: feature libraries for Gabor, Haralick, entropy, polynomial, and histograms; and charting capabilities to display line, bar, and scatter plots within Slicer.
- Compatibility with ITK version 4: Compatibility with ITK version 4 was developed and continuously maintained over the past year as ITKv4 matured. Slicer will officially switch to ITKv4 in the coming months.
Plans
At the time of this writing, 3D Slicer 4.1 is being finalized. After the release of Slicer 4.1, the Slicer development codebase will migrate to using ITK version 4. This migration will enable new image registration methods, introduce SimpleITK APIs, and introduce GPU support for image analysis algorithms. Other plans for the Computational Platform include further developments for interactive analysis methods, infrastructure for private cloud computing, and interfaces for statistical analysis.
5.2.3. Data Management Platform
Dan Marcus
Efforts in the Data Management Platform have focused on (1) developing an ergonomic user interface and internal networking logical to efficiently exchange data between Slicer and XNAT and (2) expanding Slicer's support for importing from and exporting to local DICOM Objects and networked DICOM PACS.
Major Developments
- User Interface: The XNAT development team is working with a web usability firm (Integrity St. Louis) to design and implement a web interface that can be deployed in Slicer 4's Qt framework for exploring data hosted in a remote XNAT data repository. Prototypes have been implemented and are currently being reviewed by stakeholders with the goal of a functioning interface by June, 2012.
- Networking logic: An alpha implementation of the infrastructure for actually exchanging data between XNAT and Slicer has been developed and revealed that significant additional work is required to handle parsing of MRML files within Slicer in the context of remotely hosted data. This work is now underway and will require several months to complete.
- Use cases: Several use cases have been identified, including projects within the Quantitative Imaging Network (QIN), to drive the development of the Slicer/XNAT integration.
- DICOM-RT and QIN support: Via DCMTK and custom classes, Slicer is being extended to support RT Plans, Images, Annotations, etc. Much of the support is being driving by the head-and-neck cancer core. Additional developments are being driven by an ongoing effort to integrate Slicer as an annotation module in the Quantitative Imaging Network (QIN).
- DICOM database and networking: Slicer DICOM support is approaching clinical quality in terms of speed of searching and IO by maintaining a database that indexes previously loaded DICOM objects. Via this database, it is no longer necessary to parse each object in order to search and/or load an entire series, study, or patient into Slicer.
Plans
The bulk of the effort in the coming year will continue to focus on (1) the user interface and networking logic for remote data management in XNAT and (2) import and export of Slicer results (i.e., entire MRML scenes) as DICOM objects. Regarding XNAT, as the initial development completes, we will turn to developing more efficient mechanisms for caching data locally and synchronizing local caches with remote repositories. Regarding importing and exporting Slicer data as DICOM objects, we are pursuing the concept of a DICOM Lollipop. In a DICOM Lollipop, a complete Slicer (MRML) Scene, with annotations, segmentations, viewing conditions, etc, can be saved as a binary payload in a standard DICOM object. In this manner, Slicer's data can be pushed/pulled from PACS for integration with and sharing across hospital workflows.
5.2.4. Community Software Process
Stephen R. Aylward
The goal of the Community Software Process effort is to provide tools and processes that make it easy for algorithms developers to contribute methods to the NA-MIC Kit, while maintaining the NA-MIC Kit's high-quality software standards.
Major Developments
This year, we have seen massive expansion and significant stabilization of the NA-MIC Kit. While these two accomplishments may seem at odds, they actually represent the planned progression of our community software processes. The specific aims and approaches we pursued to achieve these accomplishments are as follows:
- Provide a modern, stable platform: Slicer 4.0 has been released. This is a major milestone in the NA-MIC community. Slicer 4.0 represents a re-write of the majority of the slicer core to achieve stability, remove redundancy, refactor inefficiencies, and provide new pathways for growth. The most noticeable change is the conversion of the user interface to Qt - which provided speed, stability, and support from the well established Qt community.
- Provide a simple interface for algorithm developers to extend Slicer:
- Python has been adopted as the preferred scripting language, and it has been tightly integrated into Slicer 4.0. Python is a powerful scripting language with strong scientific computing support via add-on libraries such as SciPy, NumPy, and NiPy. We have tightly integrated it with Slicer so that python scripts can modify and extend the Slicer GUI, manipulate Slicer's data representations (i.e., the MRML Scene), and call other extensions in Slicer to specify novel workflows.
- Slicer Extension Manager is now the "Slicer Catalog." It is a new web-based system that builds upon the "App Store" concept that is familiar to Android and Apple users. Users can easily install, uninstall, rate, and comment on extensions. Developers can easily add new extensions, upload revisions, add screenshots, and respond to feedback from users.
- Provide access to the best algorithms: ITKv4 has been released by the Insight Software Consortium, and we have begun integrating this new version of ITK and its associated wrapping for Python (i.e., SimpleITK) into Slicer 4.0.
- Provide easy access to clinical data: Upgraded the version of the DICOM library (DCMTK) used by Slicer and provided improved DICOM RT support. We also have begun supporting Ultrasound (e.g., video) and 4D (e.g., gated CT) data in Slicer.
- Provide easy-to-create and easy-to-access documentation: We have integrated the extension writing and the documentation generation processes. The documentation created when an extension is written is now automatically ported to a web host for easier access from within and from outside of Slicer.
Plans
The planned efforts of the Community Software Processes are continuations of ongoing work:
- Provide a modern, stable platform: Slicer quality will continue to be improved via refactoring and via expanded emphasis on testing. Refactoring of the core is nearly complete and new efforts will be directed by the Algorithms team - to support their evolving needs and to inspire future research directions. Testing will evolve from code unit testing to GUI testing. Kitware's ParaView team has developed a semi-automated GUI testing system. It can record and then play-back user interactions. It can also verify that the user interface and Slicer output matches a pre-defined state. In this way, algorithm developers can more easily implement regression tests to ensure the continued operation of their modules.
- Provide a simple interface for algorithm developers to extend Slicer:
- Python: We will continue to promote Python as the preferred language for scripting in Slicer. It will be used for algorithm prototyping, parameter exploration, and workflow development and delivery. In particular, we expect future development to facilitate scripts that feature interactive algorithms running within Slicers 2D and 3D visualizations.
- Slicer Catalog: Future work will focus on extending the foundation introduced in Slicer 4.1. New developments will address hosting "extension packages" (e.g., the microscopy package or the DTI package of extensions) as well as hosting data, tutorials, videos, and other adjunct material.
- Provide access to the best algorithms: We will complete integration of ITKv4 and its associated SimpleITK (for python) into Slicer 4. Special attention will be given to ensuring that the upgrade will not disrupt the operation of existing modules.
- Provide easy access to clinical data: The OFFIS and CTK developers anticipate further updated to DCMTK and other clinical data systems used by Slicer. We will continue, in particular, to broaden and stabilize Slicer's support of DICOM, DICOM RT, Ultrasound, video, and 4D data.
5.3. NA-MIC Kit
The NA-MIC Kit is designed to accelerate the pace of research and transition to clinical evaluation providing (a) a flexible yet stable execution and visualization engine with strong support for clinical data (Slicer), (b) tools for extending that platform and sharing those extensions with others, and (c) community-developed analysis modules can be rapidly deployed to clinical researchers and the broader community via Slicer.
5.3.1. Expansion
Stephen R. Aylward
Progress
- Package Manager
- Updates to ITK, VTK, Qt, CMake, DCMTK, ...
- CTK
- GUI Testing
Plans
5.3.2. Release
Stephen R. Aylward