Difference between revisions of "Projects:dtistatisticsfibers"
| (One intermediate revision by the same user not shown) | |||
| Line 6: | Line 6: | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * Utah: Anuja Sharma, Sylvain Gouttard, Guido Gerig | + | * Utah Algorithms: Anuja Sharma, Sylvain Gouttard, Guido Gerig |
| − | + | * UNC Algorithms: Jean-Baptiste Berger, Benjamin Yvernault, Clement Vachet, Yundi Shi, Aditya Gupta, Martin Styner | |
<h3>Objective</h3> | <h3>Objective</h3> | ||
| Line 79: | Line 79: | ||
| + | <h3>Updates (August 2012)</h3> | ||
| + | |||
| + | A revised version of the command line tool has been incorporated in a GUI based tool [http://www.nitrc.org/projects/dti_tract_stat DTIAtlasFiberAnalyzer] available via [http://www.nitrc.org NITRC]. For details on the complete pipeline for DTI processing which makes use of this tool's functionality, please refer to the [http://www.na-mic.org/Wiki/index.php/Projects:AtlasBasedDTIFiberAnalyzerFramework UNC Algorithm page]. | ||
| + | <h3> References </h3> | ||
| + | * Casey B. Goodlett, P. Thomas Fletcher, John H. Gilmore, Guido Gerig. Group Analysis of DTI Fiber Tract Statistics with Application to Neurodevelopment. NeuroImage 45 (1) Supp. 1, 2009. p. S133-S142. | ||
[[Category:Diffusion MRI]] | [[Category:Diffusion MRI]] | ||
Latest revision as of 10:21, 19 August 2012
Home < Projects:dtistatisticsfibersStatistics on DTI data
Back to NA-MIC Collaborations, Utah2 Algorithms, MIT Algorithms, UNC Algorithms
DTI Fiber Tract statistics
Key Investigators
- Utah Algorithms: Anuja Sharma, Sylvain Gouttard, Guido Gerig
- UNC Algorithms: Jean-Baptiste Berger, Benjamin Yvernault, Clement Vachet, Yundi Shi, Aditya Gupta, Martin Styner
Objective
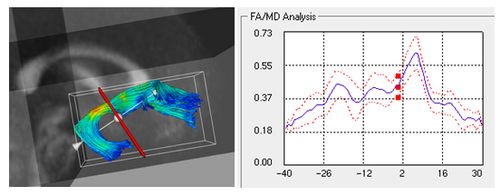
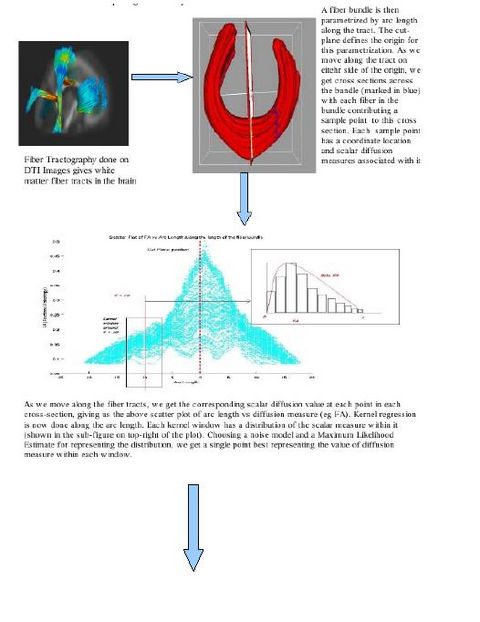
The main aim is to provide statistical information of white matter diffusion associated to fiber tracts, and therefore complements geometric information obtained by tractography with white matter integrity measurements. The tool calculates statistics on parametrized fiber tract data, represented as sets of streamlines. Using arc-length parametrization of streamline bundles, we calculate statistical parameters within cross-sections that are swept along bundles. Kernel regression along tracts provides an aperture function to include diffusion measurements of a spatial neighborhood. Diffusion invariants such as fractional anisotropy (FA), mean diffusivity (MD), Frobenius norm (FRO), axial diffusivity (AD), and transversal diffusivity (RD) are processed.
The long term goal is to use the quantitative diffusion information to understand changes of white matter structure with age, gender or specific disease processes.
Approach, Plan
We work with DTI data and follow the complete pipeline motivated by Casey Goodlett's work. (The pipeline includes unbiased non-rigid registration of a population, followed by fiber tractography applied to the average atlas and mapping the atlas geometry back to individual subjects to get the individual DTI data.) These individual subject fiber tracts are the input for the command line tool we are working on.
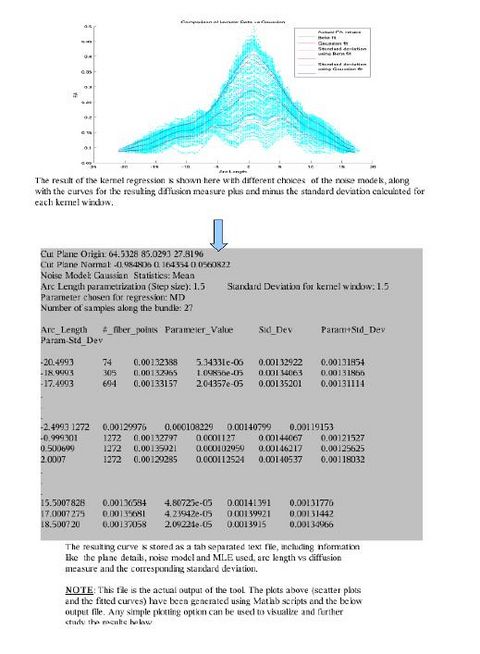
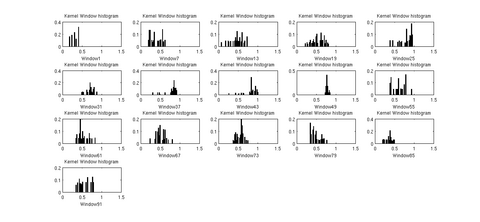
In the tool, various tract-oriented scalar diffusion measures are treated as a continuous function of fiber arc-length. To analyze the trend along the fiber tract, the command line tool performs kernel regression on this data. The idea is to try out different noise models and maximum likelihood estimates within kernel windows, such that they best represent the data and are robust to noise and Partial Volume effect.
Casey Goodlett's functional data analysis pipeline is then applied to this data. Here, multivariate hypothesis test is used to test for differences between populations and see if we get statistically significant results.
Progress
The first version of the command line tool is ready for upload into NITRC. It provides the flexibility to choose the scalar diffusion measure to be tested; a choice between Gaussian and Beta noise models and Mean, median or mode as MLE. It also incorporates several visualization options which help in analyzing the best noise models and the best representative statistics to explain the distribution of DTI data along the fiber tract. We are also working on integrating the tool into the Slicer environment. The tool can now work with UNC/UTAH .fib file format as well as the more popular VTK poly data format. The output is a csv file which can easily be used for further analysis and visualizations. The tool needs a cut-plane to define a reference origin along the fiber tract's length. We now have the option of the user visually choosing a reference plane for a fiber tract (using Fiber Viewer or a similar software) or an auto generation of a reference plane cutting the fiber bundle approximately in the middle.
The features available in the tool currently, its use and input / output formats and other relevant details are provided in the first draft of the documentation. (PDF). The document is comprehensive and also includes the concepts and the algorithm behind the tool, limitations of this approach and various visual results. The tool is still a work in progress. More features will be added to it, specially more options to plot and visualize the results. The tool will also replace the command line interface that Fiber Viewer tool currently provides to do similar (though very limited) tasks.
We are currently using the tool and the complete pipeline to study population differences longitudinally, in Autism and Cocaine related studies.
Images
Visually understanding the command line tool and the complete pipeline
More intuitive visual images and results
Updates (August 2012)
A revised version of the command line tool has been incorporated in a GUI based tool DTIAtlasFiberAnalyzer available via NITRC. For details on the complete pipeline for DTI processing which makes use of this tool's functionality, please refer to the UNC Algorithm page.
References
- Casey B. Goodlett, P. Thomas Fletcher, John H. Gilmore, Guido Gerig. Group Analysis of DTI Fiber Tract Statistics with Application to Neurodevelopment. NeuroImage 45 (1) Supp. 1, 2009. p. S133-S142.