Difference between revisions of "2013 Project Week:US IGT"
From NAMIC Wiki
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
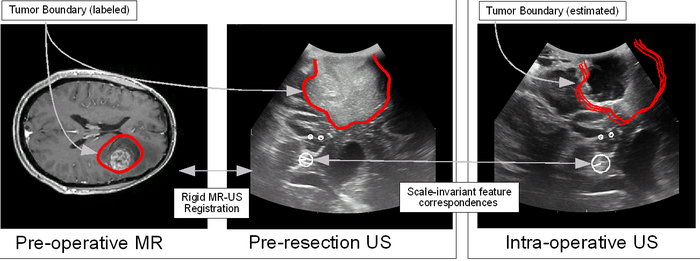
| − | [[File:US_Registration.png |700px |Ultrasound Guidance for Neurosurgery]] | + | [[File:US_Registration.png |700px |Ultrasound Guidance for Neurosurgery]] |
| + | |||
| + | |||
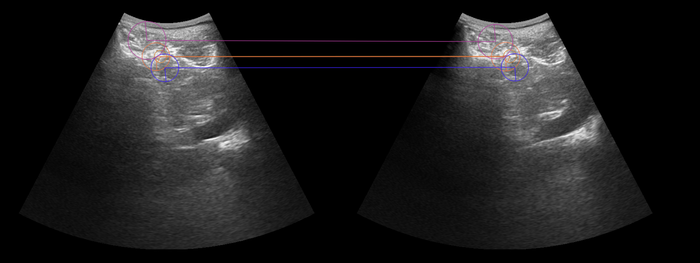
| + | Example Result: Kidney Data | ||
| + | [[Image:Animation-namic.gif |700px |Preliminary Results: Features correspondences, Kidney Ultrasound Sweeps]] | ||
| + | |||
==Key Investigators== | ==Key Investigators== | ||
Revision as of 07:48, 11 January 2013
Home < 2013 Project Week:US IGT
Key Investigators
- Matthew Toews, BWH - Harvard Medical School
- William Wells, BWH - Harvard Medical School
- Stephen Aylward, Kitware
- Tamas Ungi, Queens University
Project Description
Objective
- Develop a system to use calibrated, tracked ultrasound (US) to intra-operatively track and visualize pre-operatively defined neurosurgical targets.
- Challenges: 2D-3D registration, intra-operative deformation and missing tissue, inter-modal image registration.
Approach, Plan
- Data preparation: US image and probe tracking information (PLUS system).
- Code development: pre-op to intra-op image matching using local image features.
Progress
Developed and tested code for:
- Input of PLUS US data
- Computation of local feature correspondences between different US sweeps