Difference between revisions of "2013 Project Week:InvestigatingRoleOfOpenSourceInTranslationalResearch"
From NAMIC Wiki
(Created page with '__NOTOC__ <gallery> Image:PW-SLC2013.png|Projects List Image:yourimagehere.png| Image description </gallery> ==Key Investigators== * Junic…') |
|||
| (33 intermediate revisions by 2 users not shown) | |||
| Line 7: | Line 7: | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * Miki Kumekawa, Chiba University |
| − | |||
==Project Description== | ==Project Description== | ||
| Line 15: | Line 14: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | * | + | *Objective |
| − | ** | + | **The goal of this project week is to identify the role of open source in translational research. |
| − | *** | + | ***1 Investigating process of TR |
| − | *** | + | ***2 Investigating feature of open source (technical & social) |
| − | |||
</div> | </div> | ||
| Line 25: | Line 23: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
*Approach | *Approach | ||
| − | ** | + | **Method : Interview with 5 researchers of Slicer/software or medical device (15-30min) |
| − | ** | + | **Items : Background / Process of TR / About Open Source |
| − | *** | + | **Analysis1: structural analysis |
| − | *** | + | ***1-1 transcript data |
| + | ***1-2 coding | ||
| + | ***1-3 categolizing | ||
| + | **Analysis2: contextual analysis | ||
| + | ***2-1 transcript data | ||
| + | ***2-2 coding | ||
| + | ***2-3 categolizing(tool/human/action/conversation) | ||
| + | |||
| + | ---- | ||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | *Elements (hypothesis) |
| + | **users needs = problem | ||
| + | **technical seeds | ||
| + | |||
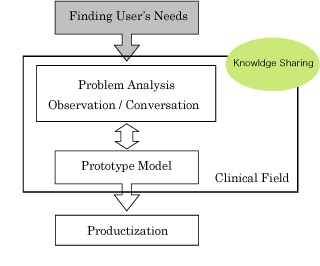
| + | *Process of Translational Research | ||
| + | [[File:flow.jpg]] | ||
| + | *Type of Needs | ||
| + | [[File:1.jpg]] | ||
| + | |||
| + | *Role of Open Source in Translational Research | ||
| + | **Extracting the user's needs directly | ||
| + | **Technical Development by users | ||
| + | **The advertisement for company | ||
| + | |||
| + | |||
| + | |||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 17:17, 11 January 2013
Home < 2013 Project Week:InvestigatingRoleOfOpenSourceInTranslationalResearch- Yourimagehere.png
Image description
Key Investigators
- Miki Kumekawa, Chiba University
Project Description
Objective
- Objective
- The goal of this project week is to identify the role of open source in translational research.
- 1 Investigating process of TR
- 2 Investigating feature of open source (technical & social)
- The goal of this project week is to identify the role of open source in translational research.
Approach, Plan
- Approach
- Method : Interview with 5 researchers of Slicer/software or medical device (15-30min)
- Items : Background / Process of TR / About Open Source
- Analysis1: structural analysis
- 1-1 transcript data
- 1-2 coding
- 1-3 categolizing
- Analysis2: contextual analysis
- 2-1 transcript data
- 2-2 coding
- 2-3 categolizing(tool/human/action/conversation)