Difference between revisions of "Projects:VentricleSegmentation"
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[Algorithm: | + | Back to [[Algorithm:Stony Brook|Stony Brook University Algorithms]] |
__NOTOC__ | __NOTOC__ | ||
| Line 10: | Line 10: | ||
surface extracted based on the left ventricle segmentation result. | surface extracted based on the left ventricle segmentation result. | ||
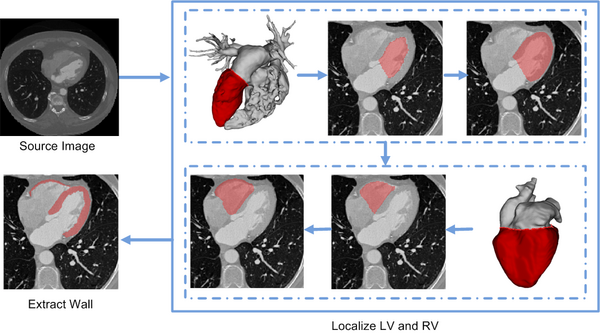
| − | *[[File:FlowChartLRV.png| | + | *[[File:FlowChartLRV.png|600px]] |
Flowchart of the ventricles segmentation framework. | Flowchart of the ventricles segmentation framework. | ||
= Results = | = Results = | ||
| + | The proposed method has been tested using 30 human and 12 pig cardiac CT images. Examples of segmentation for human and pig data are shown below. | ||
| + | |||
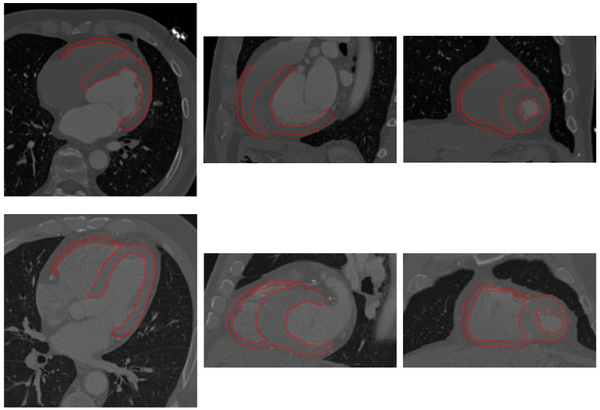
*[[File:LRVWallShapeVar.png|600px]] | *[[File:LRVWallShapeVar.png|600px]] | ||
Myocardium segmentation results of human data with significantly different heart shapes. | Myocardium segmentation results of human data with significantly different heart shapes. | ||
| Line 19: | Line 21: | ||
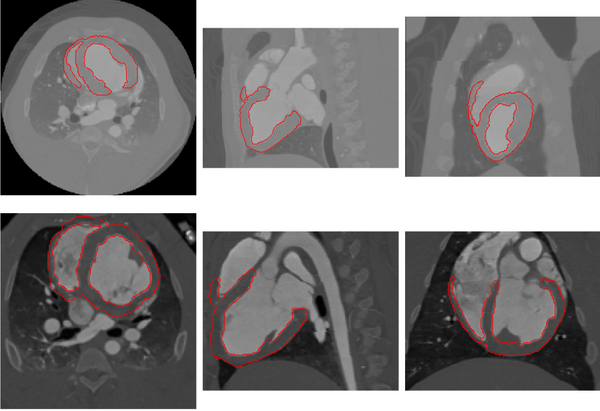
*[[File:LRVWallVolVar.png|600px]] | *[[File:LRVWallVolVar.png|600px]] | ||
Myocardium segmentation results of pig data with different volume coverages. | Myocardium segmentation results of pig data with different volume coverages. | ||
| + | |||
| + | = Key Investigators = | ||
| + | *Georgia Tech: Liangjia Zhu and Anthony Yezzi | ||
| + | *BWH: Yi Gao | ||
| + | *Boston University: Allen Tannenbaum | ||
| + | |||
| + | |||
| + | = Publication = | ||
| + | L. Zhu, Y. Gao, V. Appia, A. Yezzi, C. Arepalli , A. Stillman, and A. Tannenbaum. Automatic Extraction of the Myocardial Wall from CT Images using Shape Segmentation and Variational Region Growing, IEEE Transaction on Biomedical Engineering(TBME), vol 60, no, 10, 2013. | ||
Latest revision as of 01:57, 5 June 2014
Home < Projects:VentricleSegmentationBack to Stony Brook University Algorithms
Ventricles Segmentation
Extracting the myocardial wall of the left (LV) and right (RV) ventricles are important steps in the diagnosis of cardiac diseases. In this paper, we we propose an method for automatically extracting the ventricles from cardiac CT images, which integrates region growing with shape segmentation in a natural way. In this framework, the shape segmentation provides seed regions for region growing while the latter reconstructs a heart surface for shape decomposition.
Description
In the method, the left and right ventricles are located sequentially, in which each ventricle is detected by first identifying the endocardial surface and then segmenting the epicardial surface. To this end, the endocardial surfaces are localized using their geometric features obtained on-line from a CT image. After that, a variational region-growing model is employed to extract the epicaridal surfaces of the ventricles. In particular, the location of the endocardial surface of the left ventricle is determined using an active contour model on the blood-pool surface constructed via thresholding. To localize the right ventricle, the active contour model is performed on a heart surface extracted based on the left ventricle segmentation result.
Flowchart of the ventricles segmentation framework.
Results
The proposed method has been tested using 30 human and 12 pig cardiac CT images. Examples of segmentation for human and pig data are shown below.
Myocardium segmentation results of human data with significantly different heart shapes.
Myocardium segmentation results of pig data with different volume coverages.
Key Investigators
- Georgia Tech: Liangjia Zhu and Anthony Yezzi
- BWH: Yi Gao
- Boston University: Allen Tannenbaum
Publication
L. Zhu, Y. Gao, V. Appia, A. Yezzi, C. Arepalli , A. Stillman, and A. Tannenbaum. Automatic Extraction of the Myocardial Wall from CT Images using Shape Segmentation and Variational Region Growing, IEEE Transaction on Biomedical Engineering(TBME), vol 60, no, 10, 2013.