Difference between revisions of "2014 Summer Project Week:Slicer Murin Shape Analysis"
From NAMIC Wiki
(Created page with '==Project Description== <div style="margin: 20px;"> <div style="width: 27%; float: left; padding-right: 3%;"> Procrustes based shape analyses are a very establish set of geometr…') |
|||
| (33 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | ==Key Investigators== | ||
| + | * Murat Maga (Seattle Children's Research Institute & University of Washington Dept. of Pediatrics) | ||
| + | * Ryan Young (Seattle Children's Research Institute) | ||
| + | |||
| + | |||
==Project Description== | ==Project Description== | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| − | + | ||
| − | + | <li>Research: Changes in development due to [http://www.cdc.gov/ncbddd/fasd/index.html Fetal Alcohol Exposure] and how this affects the development of the craniofacial complex. | |
| − | We | + | <ul> |
| + | <li> Face is the major diagnostic feature to identify | ||
| + | <li> Brain and the CNS are affected primarily. | ||
| + | <li> What's the earliest time we begin to detect changes in the face? | ||
| + | <li> How does the brain volumes (and gross morphology) relate to changes in the face? | ||
| + | </ul> | ||
| + | |||
| + | <li> Modalities: <b> Optical Projection Tomography</b> [[File:Sample OPT Mouse embryo.zip]] <br> | ||
| + | <B> Micro Computed Tomography </b> [[File:Stained registered sample mCT.zip]] <br> | ||
| + | [[Image:OPT Crossection.PNG|100px]] [[Image:Registered mCT scans.png|100px]] | ||
| + | <li> Shape Analysis | ||
| + | <ul> | ||
| + | <li>We use landmarks to identify the anatomical regions across our developmental series of fetal samples. | ||
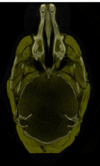
| + | <li>We want to be able segment brains from about 600 volumes and do a coupled analysis of facial and brain phenotypes. <br> | ||
| + | [[Image:Fetus variation picture.PNG|400px]] | ||
| + | </ul> | ||
| + | |||
| + | <li> Challenges in Slicer with our datasets due to small voxel sizes (6-35 micron). Specifically visualization, recording coordinates of anatomical landmarks, segmentation and registration. ([[File:Project week question.txt]]) | ||
| + | |||
| + | <li> Goals for Project Week: | ||
| + | <ul> | ||
| + | <li> Meet the community and learn from them! | ||
| + | <li> Raise awareness about issues in using Slicer in high-resolution small animal imaging. | ||
| + | <li> Implement the landmark based Procrustes Analysis in Slicer | ||
| + | </ul> | ||
| + | |||
| + | |||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | Create a GPA/PCA shape analysis and visualization module for Slicer. | + | *Create a GPA/PCA shape analysis and visualization module for Slicer. |
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
<ul> | <ul> | ||
| − | <li> | + | <li>Implement GPA/PCA shape analysis in python</li> |
| − | <li> | + | <li>Provide an interactive tool to visualize the decomposition along the principle components of shape variation using thin plate splines.<br> |
| + | [[Image:TPS.png|400px]])</li> | ||
| − | <li>Ability to create semi-landmarks to increase | + | <li> <b>Ability to create semi-landmarks to increase coverage in regions where anatomial landmarks are sparse. </b> |
<ul> | <ul> | ||
| − | <li>User will | + | <li><b>User will a uniformly sampled point cloud by entering the number of semi-landmarks. Existing “hard” landmarks will be used for their distribution. This will serve as the template to be transferred to all remaining volumes (atlas) |
<li>The template will be transferred to a new surface. Existing “hard” landmarks will allow for correspondence. The transferred points will then be moved along the surface of the volume by optimizing the bending energy function. | <li>The template will be transferred to a new surface. Existing “hard” landmarks will allow for correspondence. The transferred points will then be moved along the surface of the volume by optimizing the bending energy function. | ||
| − | <li>The coordinates of the slid landmarks will be saved into a new fiducial list, from which the GPA analysis can be conducted. | + | <li>The coordinates of the slid landmarks will be saved into a new fiducial list, from which the GPA analysis can be conducted.</b> |
| + | <li>These should be accomplished on volume, not surface meshes derived from scans. | ||
</ul> | </ul> | ||
</ul> | </ul> | ||
| Line 26: | Line 59: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
<ul> | <ul> | ||
| − | <li> Generalized Procrustes Alignment</li> | + | <li> Generalized Procrustes Alignment (implemented)</li> |
| − | <li> Principal Component and Singular Value Decomposition of the Procrustes aligned coordinates</li> | + | <li> Principal Component and Singular Value Decomposition of the Procrustes aligned coordinates (implemented)</li> |
| − | <li> Thin Plate Spline visualization of the shape variables from PCA and/or SVD ( | + | <li> Thin Plate Spline visualization of the shape variables from PCA and/or SVD (implemented).</li> |
| + | <li> Transfering and sliding a template of semi-landmarks to the target volume (in progress) | ||
</ul> | </ul> | ||
| − | + | ||
| + | [[File:PowerPoint.pdf|Intro Power Point]] | ||
</div> | </div> | ||
Latest revision as of 15:46, 24 June 2014
Home < 2014 Summer Project Week:Slicer Murin Shape AnalysisKey Investigators
- Murat Maga (Seattle Children's Research Institute & University of Washington Dept. of Pediatrics)
- Ryan Young (Seattle Children's Research Institute)
Project Description
- Face is the major diagnostic feature to identify
- Brain and the CNS are affected primarily.
- What's the earliest time we begin to detect changes in the face?
- How does the brain volumes (and gross morphology) relate to changes in the face?
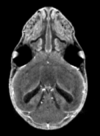
Micro Computed Tomography File:Stained registered sample mCT.zip
- Meet the community and learn from them!
- Raise awareness about issues in using Slicer in high-resolution small animal imaging.
- Implement the landmark based Procrustes Analysis in Slicer
Objective
- Create a GPA/PCA shape analysis and visualization module for Slicer.
Approach, Plan
- Implement GPA/PCA shape analysis in python
- Provide an interactive tool to visualize the decomposition along the principle components of shape variation using thin plate splines.
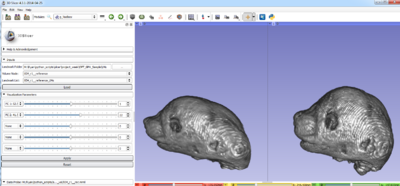 )
) - Ability to create semi-landmarks to increase coverage in regions where anatomial landmarks are sparse.
- User will a uniformly sampled point cloud by entering the number of semi-landmarks. Existing “hard” landmarks will be used for their distribution. This will serve as the template to be transferred to all remaining volumes (atlas)
- The template will be transferred to a new surface. Existing “hard” landmarks will allow for correspondence. The transferred points will then be moved along the surface of the volume by optimizing the bending energy function.
- The coordinates of the slid landmarks will be saved into a new fiducial list, from which the GPA analysis can be conducted.
- These should be accomplished on volume, not surface meshes derived from scans.
Progress
- Generalized Procrustes Alignment (implemented)
- Principal Component and Singular Value Decomposition of the Procrustes aligned coordinates (implemented)
- Thin Plate Spline visualization of the shape variables from PCA and/or SVD (implemented).
- Transfering and sliding a template of semi-landmarks to the target volume (in progress)
