Difference between revisions of "2014 Summer Project Week:Slicer LDDMM Shape Analysis"
From NAMIC Wiki
Saurabhjain (talk | contribs) |
Saurabhjain (talk | contribs) |
||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
| + | <gallery> | ||
| + | Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | ||
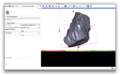
Image:LDDMM-Plugin-Screen-Shot.png | Image:LDDMM-Plugin-Screen-Shot.png | ||
| + | Image:ScreenShot_2014-06-26.png | ||
</gallery> | </gallery> | ||
| Line 6: | Line 10: | ||
* Isomics: Steve Pieper | * Isomics: Steve Pieper | ||
* Iowa: Hans Johnson | * Iowa: Hans Johnson | ||
| − | * | + | * Iowa: Regina Young |
| Line 14: | Line 18: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | Develop a Slicer interface to | + | Develop a Slicer interface to various remote processing services |
| − | for | + | for shape analysis of human brain sub-cortical structures offered by |
| − | Center for Imaging Science (CIS) through our | + | Center for Imaging Science (CIS) through our portal. |
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| + | * The pipeline accepts surfaces stored in the BYU format, or binary masks stored in the ANALYZE format. The processing is done on one of the clusters available to CIS, and upon completion the user receives an e-mail with a link to the results stored on the ftp site. | ||
| + | |||
| + | * At the previous project week, we created a simple interface, which accepts the directory location, and e-mail address (which must be registered with http://www.MRIStudio.org), and sends a template estimation job if the directory contains .byu files. To receive the estimated template, the user copies the md5 checksum from the e-mail, which reports the job completion, to the md5 box in slicer and hit receive. | ||
| + | |||
* The template estimation is performed using a variant of the approach described in: | * The template estimation is performed using a variant of the approach described in: | ||
| Line 27: | Line 35: | ||
vol. 2010, Article ID 974957. | vol. 2010, Article ID 974957. | ||
| − | * | + | * We now plan to create interfaces to other services including LDDMM surface matching, and geodesic time series matching and template estimation for longitudinal data. |
| − | * | + | * In addition, we plan to make the interface more user friendly, which includes support for multiple segmentation labels stored as NiFTI files for data such as Han's (see 'Progress' tab), getting input from Slicer scenes, and importing the processed results to scenes. |
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | |||
| − | |||
| − | |||
* Hans shared datasets for testing: | * Hans shared datasets for testing: | ||
https://www.dropbox.com/sh/5w2ls8b4j9ihpze/51IkhaXbTJ | https://www.dropbox.com/sh/5w2ls8b4j9ihpze/51IkhaXbTJ | ||
| + | * Added interface for LDDMM mappings, in addition to the template estimation interface developed last time. | ||
| + | * Regina used the plugin to send a test job. | ||
| + | * Made the interface a bit more user friendly. | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 03:31, 27 June 2014
Home < 2014 Summer Project Week:Slicer LDDMM Shape AnalysisKey Investigators
- JHU: Saurabh Jain
- Isomics: Steve Pieper
- Iowa: Hans Johnson
- Iowa: Regina Young
Project Description
Objective
Develop a Slicer interface to various remote processing services for shape analysis of human brain sub-cortical structures offered by Center for Imaging Science (CIS) through our portal.
Approach, Plan
- The pipeline accepts surfaces stored in the BYU format, or binary masks stored in the ANALYZE format. The processing is done on one of the clusters available to CIS, and upon completion the user receives an e-mail with a link to the results stored on the ftp site.
- At the previous project week, we created a simple interface, which accepts the directory location, and e-mail address (which must be registered with http://www.MRIStudio.org), and sends a template estimation job if the directory contains .byu files. To receive the estimated template, the user copies the md5 checksum from the e-mail, which reports the job completion, to the md5 box in slicer and hit receive.
- The template estimation is performed using a variant of the approach described in:
J. Ma, M. I. Miller, and L. Younes, “A Bayesian Generative Model for Surface Template Estimation,” Intl. J. Bio. Img., vol. 2010, Article ID 974957.
- We now plan to create interfaces to other services including LDDMM surface matching, and geodesic time series matching and template estimation for longitudinal data.
- In addition, we plan to make the interface more user friendly, which includes support for multiple segmentation labels stored as NiFTI files for data such as Han's (see 'Progress' tab), getting input from Slicer scenes, and importing the processed results to scenes.
Progress
- Hans shared datasets for testing:
https://www.dropbox.com/sh/5w2ls8b4j9ihpze/51IkhaXbTJ
- Added interface for LDDMM mappings, in addition to the template estimation interface developed last time.
- Regina used the plugin to send a test job.
- Made the interface a bit more user friendly.