Difference between revisions of "2014 Summer Project Week:Cardiac-Congenital"
From NAMIC Wiki
(Created page with '__NOTOC__ <gallery> Image:PW-MIT2014.png|Projects List </gallery> ==Key Investigators== * Danielle Pace, MIT * Adrian Dalca, MIT * Polina G…') |
|||
| (4 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | ||
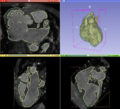
| + | Image:CongenitalHeartModelsSlicer.png|Results of patch-based segmentation | ||
| + | Image:CongenitalHeartModels.png|Patient-specific heart model | ||
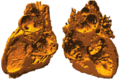
| + | Image:CongenitalHeartModels2.png|Printed model | ||
| + | |||
</gallery> | </gallery> | ||
| Line 10: | Line 14: | ||
==Project Description== | ==Project Description== | ||
| + | |||
| + | This project involves semi-automatic segmentation of gated 3D magnetic resonance images of hearts with congenital heart defects. Our aim is to create patient-specific heart models for surgical planning, which can be viewed either graphically on a computer, or with a 3D printer to create a physical model for surgeons. | ||
| + | |||
| + | We have had initial success in significantly reducing segmentation time with the following pipeline: | ||
| + | 1) User manually segments ~10 axial slices | ||
| + | 2) Segment the remaining slices using patch-based majority voting. | ||
| + | |||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | * | + | * Continue testing the patch based methods on an additional five datasets. |
| + | * Address main remaining challenge: segmenting thin interior heart walls. | ||
| + | |||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | * | + | * Manually segment ~10 axial slices for each of the five additional datasets. |
| + | * Explore remaining parameters for the patch-based segmentation: e.g. weighted voting, varying k in k-nearest neighbors patch lookup | ||
| + | |||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | * Did manual initial segmentations and and ran patch-based segmentation on additional datasets. |
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 04:32, 27 June 2014
Home < 2014 Summer Project Week:Cardiac-CongenitalKey Investigators
- Danielle Pace, MIT
- Adrian Dalca, MIT
- Polina Golland, MIT
Project Description
This project involves semi-automatic segmentation of gated 3D magnetic resonance images of hearts with congenital heart defects. Our aim is to create patient-specific heart models for surgical planning, which can be viewed either graphically on a computer, or with a 3D printer to create a physical model for surgeons.
We have had initial success in significantly reducing segmentation time with the following pipeline: 1) User manually segments ~10 axial slices 2) Segment the remaining slices using patch-based majority voting.
Objective
- Continue testing the patch based methods on an additional five datasets.
- Address main remaining challenge: segmenting thin interior heart walls.
Approach, Plan
- Manually segment ~10 axial slices for each of the five additional datasets.
- Explore remaining parameters for the patch-based segmentation: e.g. weighted voting, varying k in k-nearest neighbors patch lookup
Progress
- Did manual initial segmentations and and ran patch-based segmentation on additional datasets.