Difference between revisions of "2014 Summer Project Week:DRAMMS Slicer"
Ouyangming (talk | contribs) |
|||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | ||
Image:Dramms-logo.png|[[http://www.cbica.upenn.edu/sbia/software/dramms/ |DRAMMS Website]] | Image:Dramms-logo.png|[[http://www.cbica.upenn.edu/sbia/software/dramms/ |DRAMMS Website]] | ||
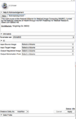
| + | Image:DRAMMS-mockup-Screen_Shot_2014-06-27_at_8.58.25_AM.png | DRAMMS UI build with python script | ||
</gallery> | </gallery> | ||
| Line 16: | Line 17: | ||
==Project Description== | ==Project Description== | ||
| − | [[http://www.cbica.upenn.edu/sbia/software/dramms/ DRAMMS]] is a general-purpose, fully-automated, deformable image registration algorithm and software package. It was designed for 2D-to-2D and 3D-to-3D cross-subject, longitudinal, multi-modal registration tasks, and has been recently extended for spatial-temporal (4D) | + | [[http://www.cbica.upenn.edu/sbia/software/dramms/ DRAMMS]] is a general-purpose, fully-automated, deformable image registration algorithm and software package. It was designed for '''2D-to-2D''' and '''3D-to-3D''' cross-subject, longitudinal, multi-modal registration tasks, and has been recently extended for '''spatial-temporal (4D)''' registration and '''population-wise''' registration. Some typical applications include: |
| + | |||
| + | * atlas construction in a population (infants, neonates, adults, ...); | ||
| + | * multi-atlas based structure segmentation (brain, prostate, ...); | ||
| + | * multi-modal registration (we had some success in histology-MRI registration of the prostate and mouse brain, looking for data for validation); | ||
| + | * registration under a partial loss of correspondences(such as tumor/lesion-bearing images to normal-appearance images); | ||
| + | * quantification of 4D tumor changes; | ||
| + | * study of early-life longitudinal neuro-deveolopment; | ||
| + | * maintenance of 4D segmentation consistency. | ||
| + | |||
| + | |||
| + | Several evaluation studies comparing DRAMMS with many state-of-the-art registration algorithms are also being documented for brain [[http://ieeexplore.ieee.org/xpl/articleDetails.jsp?tp=&arnumber=6834815&sortType%3Dasc_p_Sequence%26filter%3DAND%28p_IS_Number%3A4359023%29 DRAMMS_in_Brain_Registration]], cardiac [[http://link.springer.com/chapter/10.1007%2F978-3-642-31340-0_22 DRAMMS_in_Cardiac_Registration]], breast [[https://drive.google.com/file/d/0B1RDNI9g7RfrTHhpU21semdYYjA/edit?usp=sharing DRAMMS_in_Breast_Registration]] registration tasks. | ||
| + | |||
| + | |||
| + | The integration of DRAMMS into Slicer is expected to bring more diverse deformable registration options into Slicer, especially an option to deal with registration of both brain and non-brain structures, and to deal with registration under a partial loss of correspondences. | ||
| Line 30: | Line 45: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | * Decided that a first step would be to try running existing dramms executable through a wrapper script |
| + | ** This will let people try the tool in slicer without extensive rework | ||
| + | ** Prototype version: https://github.com/ouyangming/DRAMMS_in_Slicer/pull/1 | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 38: | Line 55: | ||
* [http://www.sciencedirect.com/science/article/pii/S1361841510000940 DRAMMS Algorithm] | * [http://www.sciencedirect.com/science/article/pii/S1361841510000940 DRAMMS Algorithm] | ||
| − | * [http://www.cbica.upenn.edu/sbia/software/dramms/ DRAMMS Software] | + | * [http://www.cbica.upenn.edu/sbia/software/dramms/ DRAMMS Software Website] |
| + | |||
| + | * [http://www.cbica.upenn.edu/sbia/software/dramms/_downloads/DRAMMS_Software_Manual.pdf DRAMMS Software Manual (pdf)] | ||
* [http://ieeexplore.ieee.org/xpl/articleDetails.jsp?tp=&arnumber=6834815&sortType%3Dasc_p_Sequence%26filter%3DAND%28p_IS_Number%3A4359023%29 Evaluation of DRAMMS in Across-Subject Brain Registration] | * [http://ieeexplore.ieee.org/xpl/articleDetails.jsp?tp=&arnumber=6834815&sortType%3Dasc_p_Sequence%26filter%3DAND%28p_IS_Number%3A4359023%29 Evaluation of DRAMMS in Across-Subject Brain Registration] | ||
| Line 44: | Line 63: | ||
* [http://link.springer.com/chapter/10.1007%2F978-3-642-31340-0_22 Evaluation of DRAMMS in Across-Subject Cardiac Registration] | * [http://link.springer.com/chapter/10.1007%2F978-3-642-31340-0_22 Evaluation of DRAMMS in Across-Subject Cardiac Registration] | ||
| − | * [Evaluation of DRAMMS in Longitudinal Breast Registration] | + | * [https://drive.google.com/file/d/0B1RDNI9g7RfrTHhpU21semdYYjA/edit?usp=sharing Evaluation of DRAMMS in Longitudinal Breast Registration] |
Latest revision as of 13:15, 27 June 2014
Home < 2014 Summer Project Week:DRAMMS SlicerKey Investigators
- MGH: Yangming Ou, Randy Gollub, Jayashree Kalpathy-Cramer
- Isomics: Steve Pieper
- UPenn: Christos Davatzikos
- BWH: Andriy Fedorov, Tina Kapur, Ron Kikinis
Project Description
[DRAMMS] is a general-purpose, fully-automated, deformable image registration algorithm and software package. It was designed for 2D-to-2D and 3D-to-3D cross-subject, longitudinal, multi-modal registration tasks, and has been recently extended for spatial-temporal (4D) registration and population-wise registration. Some typical applications include:
- atlas construction in a population (infants, neonates, adults, ...);
- multi-atlas based structure segmentation (brain, prostate, ...);
- multi-modal registration (we had some success in histology-MRI registration of the prostate and mouse brain, looking for data for validation);
- registration under a partial loss of correspondences(such as tumor/lesion-bearing images to normal-appearance images);
- quantification of 4D tumor changes;
- study of early-life longitudinal neuro-deveolopment;
- maintenance of 4D segmentation consistency.
Several evaluation studies comparing DRAMMS with many state-of-the-art registration algorithms are also being documented for brain [DRAMMS_in_Brain_Registration], cardiac [DRAMMS_in_Cardiac_Registration], breast [DRAMMS_in_Breast_Registration] registration tasks.
The integration of DRAMMS into Slicer is expected to bring more diverse deformable registration options into Slicer, especially an option to deal with registration of both brain and non-brain structures, and to deal with registration under a partial loss of correspondences.
Objective
- To integrate the DRAMMS deformable registration software into 3DSlicer.
Approach, Plan
- Starting from the current stand-alone DRAMMS version (academic- but not commercial-friendly), figure out the necessary interface to integrate it into Slicer.
Progress
- Decided that a first step would be to try running existing dramms executable through a wrapper script
- This will let people try the tool in slicer without extensive rework
- Prototype version: https://github.com/ouyangming/DRAMMS_in_Slicer/pull/1