Difference between revisions of "2014 Summer Project Week:Multidim Data"
From NAMIC Wiki
(Created page with '__NOTOC__ <gallery> Image:PW-MIT2014.png|Projects List </gallery> ==Key Investigators== ==Project Description== <div style="margin: 20px;…') |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | ||
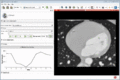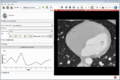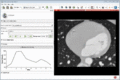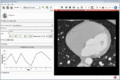
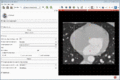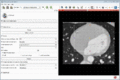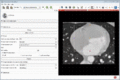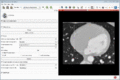
| + | Image:MultidimDataInteractivePlotting.gif|Interactive plotting of voxel intensity over time. | ||
| + | Image:MultidimDataRegistration.gif|Registration of 4D sequences and visualization of the resulting 4D transforms. | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| + | * Andras Lasso (Queen's U) | ||
| + | * Kevin Wang (Princess Margaret Cancer Center) | ||
==Project Description== | ==Project Description== | ||
| Line 11: | Line 15: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | * | + | * Ongoing work to improve the current multidim data work to support more use cases. |
| + | * Migrate part/all functionalities in MulitVolume extension into Multidim extension. | ||
| + | |||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | * | + | * Support for transform (linear and deformable) sequence data type. |
| + | * Support for label statistics for multidim data. | ||
| + | * Motion tracking module. | ||
| + | * Generic CLI support: pass multiple file names for node sequences | ||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | * Added mouse cursor position to vtkMRMLCrosshairNode (https://github.com/Slicer/Slicer/pull/147), so any module can easily observe the current mouse position in slice viewers. Other pointing devices can use it, too (LeapMotion, etc). |
| + | ** Used also by Data probe module and Transform module info window | ||
| + | * Integrated features developed by Kevin Wang: | ||
| + | ** Interactive plotting of volume intensities over time | ||
| + | ** Volume sequence registration | ||
| + | ** Multidimensional labelmap statistics | ||
</div> | </div> | ||
| + | </div> | ||
| + | |||
| + | <div style="width: 97%; float: left;"> | ||
| + | ==References== | ||
| + | * Multidim data extension source code: [https://www.assembla.com/code/slicerrt/subversion/nodes/1794/trunk/MultidimData/src] | ||
</div> | </div> | ||
Latest revision as of 14:55, 27 June 2014
Home < 2014 Summer Project Week:Multidim DataKey Investigators
- Andras Lasso (Queen's U)
- Kevin Wang (Princess Margaret Cancer Center)
Project Description
Objective
- Ongoing work to improve the current multidim data work to support more use cases.
- Migrate part/all functionalities in MulitVolume extension into Multidim extension.
Approach, Plan
- Support for transform (linear and deformable) sequence data type.
- Support for label statistics for multidim data.
- Motion tracking module.
- Generic CLI support: pass multiple file names for node sequences
Progress
- Added mouse cursor position to vtkMRMLCrosshairNode (https://github.com/Slicer/Slicer/pull/147), so any module can easily observe the current mouse position in slice viewers. Other pointing devices can use it, too (LeapMotion, etc).
- Used also by Data probe module and Transform module info window
- Integrated features developed by Kevin Wang:
- Interactive plotting of volume intensities over time
- Volume sequence registration
- Multidimensional labelmap statistics
References
- Multidim data extension source code: [1]