Difference between revisions of "T1 mapping for variable flip angle"
From NAMIC Wiki
Stevedaxiao (talk | contribs) |
|||
| Line 33: | Line 33: | ||
* [https://dblab.duhs.duke.edu/modules/QIBAcontent/index.php?id=1 QIBA T1 phantom] | * [https://dblab.duhs.duke.edu/modules/QIBAcontent/index.php?id=1 QIBA T1 phantom] | ||
* [http://europepmc.org/articles/pmc3620726 Basic equation for T1] | * [http://europepmc.org/articles/pmc3620726 Basic equation for T1] | ||
| + | * [http://slicer-devel.65872.n3.nabble.com/VTK-volume-from-numpy-Array-VTK-volume-td4033216.html how to create a new vtk volume node] | ||
| + | * [http://www.vtk.org/Wiki/index.php?title=VTK/Examples/Python/vtkWithNumpy&oldid=37957 numpy array in vtk] | ||
Revision as of 18:05, 9 January 2015
Home < T1 mapping for variable flip angleKey Investigators
- MGH: Xiao Da, Artem Mamonov, Jayashree Kalpathy-Cramer
- BWH: Andriy Fedorov
Project Description
Objective
- Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles
Approach, Plan
- Create a Slicer module for T1 mapping for multiple flip angles using Python script
- Run Python script and Freesurfer code on QIBA T1 phantom
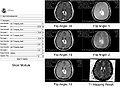
- Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles
Progress
- Compared the results using Python with Freesurfer and ground truth of T1 for QIBA data
- T1 mapping results on QIBA data using Slicer+Python are almost identical to the results using Freesurfer and comparable with the ground truth