Difference between revisions of "T1 mapping for variable flip angle"
From NAMIC Wiki
| (One intermediate revision by the same user not shown) | |||
| Line 15: | Line 15: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
* Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles | * Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles | ||
| + | * Implement T1 mapping algorithm as a Slicer extension. | ||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| Line 21: | Line 22: | ||
* Run Python script and [https://surfer.nmr.mgh.harvard.edu/fswiki/mri_ms_fitparms Freesurfer code] on [https://dblab.duhs.duke.edu/modules/QIBAcontent/index.php?id=1 QIBA T1 phantom] | * Run Python script and [https://surfer.nmr.mgh.harvard.edu/fswiki/mri_ms_fitparms Freesurfer code] on [https://dblab.duhs.duke.edu/modules/QIBAcontent/index.php?id=1 QIBA T1 phantom] | ||
* Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles | * Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles | ||
| + | * Go through Slicer tutorials and developer forum to learn how to implement Slicer extensions. | ||
| + | * Go over VTK documentation to learn how it works. | ||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
* Compared the results using Python with Freesurfer and ground truth of T1 for QIBA data | * Compared the results using Python with Freesurfer and ground truth of T1 for QIBA data | ||
| − | * T1 mapping results on QIBA data using Slicer+Python are almost identical to the results using Freesurfer and comparable with the ground truth | + | * T1 mapping results on QIBA data using Slicer+Python are almost identical to the results using Freesurfer and comparable with the ground truth. |
| + | * Implemented T1 mapping as a Slicer extension using python. The extension has GUI that as an input takes six DICOM images with corresponding flip angles and repetition time. The extension computes T1s and stores them in a new VTK volume node. | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 22:17, 10 January 2015
Home < T1 mapping for variable flip angleKey Investigators
- MGH: Xiao Da, Artem Mamonov, Jayashree Kalpathy-Cramer
- BWH: Andriy Fedorov
Project Description
Objective
- Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles
- Implement T1 mapping algorithm as a Slicer extension.
Approach, Plan
- Create a Slicer module for T1 mapping for multiple flip angles using Python script
- Run Python script and Freesurfer code on QIBA T1 phantom
- Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles
- Go through Slicer tutorials and developer forum to learn how to implement Slicer extensions.
- Go over VTK documentation to learn how it works.
Progress
- Compared the results using Python with Freesurfer and ground truth of T1 for QIBA data
- T1 mapping results on QIBA data using Slicer+Python are almost identical to the results using Freesurfer and comparable with the ground truth.
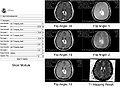
- Implemented T1 mapping as a Slicer extension using python. The extension has GUI that as an input takes six DICOM images with corresponding flip angles and repetition time. The extension computes T1s and stores them in a new VTK volume node.