Difference between revisions of "NA-MIC Brains Collaboration"
From NAMIC Wiki
| (21 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[NA-MIC_External_Collaborations]] | + | __NOTOC__ |
| + | Back to [[NA-MIC_External_Collaborations|NA-MIC External Collaborations]] | ||
==Abstract== | ==Abstract== | ||
This project is not an NCBC collaboration grant, but instead a Continued Development and Maintenance of Software grant. The intent of this application is to update the '''BRAINS''' image analysis software developed at the University of Iowa. | This project is not an NCBC collaboration grant, but instead a Continued Development and Maintenance of Software grant. The intent of this application is to update the '''BRAINS''' image analysis software developed at the University of Iowa. | ||
| − | ==Grant #== | + | ==Grant#== |
| − | + | R01NS050568 | |
==Key Personnel== | ==Key Personnel== | ||
| − | + | *University of Iowa: Vincent Magnotta, PI, Hans Johnson, Jeremy Bockholt, and Nancy Andreasen | |
| + | *NA-MIC: Steve Pieper (Isomics) | ||
| + | |||
| + | ==Funding Duration== | ||
| + | 05/15/2007-03/31/2010 | ||
==Projects== | ==Projects== | ||
| Line 17: | Line 22: | ||
*[[Support for Batch Processing]] | *[[Support for Batch Processing]] | ||
*[[Visual Review and Verification of Results]] | *[[Visual Review and Verification of Results]] | ||
| + | *[[Build GUI using Slicer3 Interface]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | === Validation of Pipeline | + | === Validation of Pipeline === |
*[[Evaluate MIND MCIC Human Phantom Sample Using Automated Pipeline]] | *[[Evaluate MIND MCIC Human Phantom Sample Using Automated Pipeline]] | ||
| − | *[[Evaluate | + | *[[Evaluate of DTI Reliability Across Sites and Platforms]] |
| − | ===Tools Developed=== | + | ===Tools Developed=== |
Here is a list of tools that are being developed as part of this project. A number of these tools have been contributed to the NITRC website. All of the tools are built upon the Slicer3 execution model and therefore will readily plug-in to Slicer3. | Here is a list of tools that are being developed as part of this project. A number of these tools have been contributed to the NITRC website. All of the tools are built upon the Slicer3 execution model and therefore will readily plug-in to Slicer3. | ||
| − | *BRAINSFit - Linear fitting of two 3D image sets using a mutual information similarity metric | + | *[[BRAINSFit]] - Linear fitting of two 3D image sets using a mutual information similarity metric. [http://www.nitrc.org/projects/multimodereg/ NITRC Link] |
| − | *BRAINSDemonsWarp - Non-linear registration of two image sets. This includes the diffeomorphic demons algorithm along with a number of useful enhancements. | + | *[[BRAINSDemonsWarp]] - Non-linear registration of two image sets. This includes the diffeomorphic demons algorithm along with a number of useful enhancements. [http://www.nitrc.org/projects/brainsdemonwarp/ NITRC Link] |
| − | *BRAINSMush - Tool to combine T1 and T2 datasets into a composite image. The resulting image is then used to perform brain extraction. | + | *[[BRAINSMush]] - Tool to combine T1 and T2 datasets into a composite image. The resulting image is then used to perform brain extraction. [http://www.nitrc.org/projects/brainsmush/ NITRC Link] |
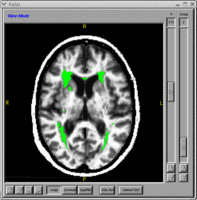
| − | * | + | *[[BRAINSCut]] - Automated segmentation of subcortical and cerebellar regions of interest using an artificial neural network. [http://www.nitrc.org/projects/brainscut/ NITRC Link] |
| − | *BRAINSTracer - Tracing tool that uses thge vtkContourWidget for defining a region of interest manually | + | *[[BRAINSTracer]] - Tracing tool that uses thge vtkContourWidget for defining a region of interest manually. [http://www.nitrc.org/projects/brainstracer/ NITRC Link] |
| − | *GTRACT - Diffusion tensor analysis and Fiber tracking tool | + | *[[GTRACT]] - Diffusion tensor analysis and Fiber tracking tool. [http://www.nitrc.org/projects/vmagnotta/ NITRC Link] |
| − | *BRAINS | + | *[[BRAINS Automated Pipeline]] - Fully automated image analysis pipeline for generation of volumetric measurements. [http://www.nitrc.org/projects/brains/ NITRC Link] |
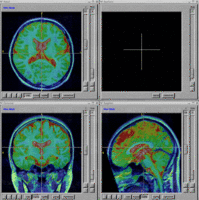
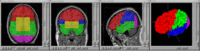
=== BRAINS Features === | === BRAINS Features === | ||
| Line 50: | Line 52: | ||
==Publications== | ==Publications== | ||
| − | + | *Powell S., Magnotta V.A., Johnson H., Jammalamadaka V.K., Pierson R., Andreasen N.C. [http://www.na-mic.org/publications/item/view/1755 Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures.] Neuroimage. 2008 Jan 1;39(1):238-47. PMID: 17904870. PMCID: PMC2253948. | |
| − | |||
==Meetings== | ==Meetings== | ||
==Resource Links== | ==Resource Links== | ||
Latest revision as of 16:57, 14 December 2016
Home < NA-MIC Brains CollaborationBack to NA-MIC External Collaborations
Abstract
This project is not an NCBC collaboration grant, but instead a Continued Development and Maintenance of Software grant. The intent of this application is to update the BRAINS image analysis software developed at the University of Iowa.
Grant#
R01NS050568
Key Personnel
- University of Iowa: Vincent Magnotta, PI, Hans Johnson, Jeremy Bockholt, and Nancy Andreasen
- NA-MIC: Steve Pieper (Isomics)
Funding Duration
05/15/2007-03/31/2010
Projects
There are three main thrusts of this application as outlined below.
Implement an Automated Brain Analysis Pipeline
- BRAINS Automated Pipeline
- Support for Batch Processing
- Visual Review and Verification of Results
- Build GUI using Slicer3 Interface
Validation of Pipeline
- Evaluate MIND MCIC Human Phantom Sample Using Automated Pipeline
- Evaluate of DTI Reliability Across Sites and Platforms
Tools Developed
Here is a list of tools that are being developed as part of this project. A number of these tools have been contributed to the NITRC website. All of the tools are built upon the Slicer3 execution model and therefore will readily plug-in to Slicer3.
- BRAINSFit - Linear fitting of two 3D image sets using a mutual information similarity metric. NITRC Link
- BRAINSDemonsWarp - Non-linear registration of two image sets. This includes the diffeomorphic demons algorithm along with a number of useful enhancements. NITRC Link
- BRAINSMush - Tool to combine T1 and T2 datasets into a composite image. The resulting image is then used to perform brain extraction. NITRC Link
- BRAINSCut - Automated segmentation of subcortical and cerebellar regions of interest using an artificial neural network. NITRC Link
- BRAINSTracer - Tracing tool that uses thge vtkContourWidget for defining a region of interest manually. NITRC Link
- GTRACT - Diffusion tensor analysis and Fiber tracking tool. NITRC Link
- BRAINS Automated Pipeline - Fully automated image analysis pipeline for generation of volumetric measurements. NITRC Link
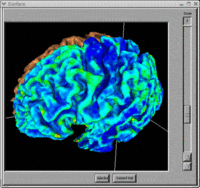
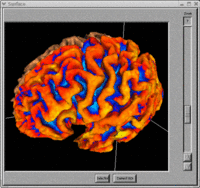
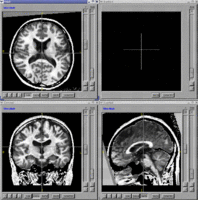
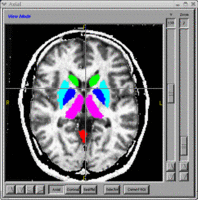
BRAINS Features
- BRAINS Gallery
Publications
- Powell S., Magnotta V.A., Johnson H., Jammalamadaka V.K., Pierson R., Andreasen N.C. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. Neuroimage. 2008 Jan 1;39(1):238-47. PMID: 17904870. PMCID: PMC2253948.