Difference between revisions of "Project Week 25/NeedleSegmentation"
From NAMIC Wiki
| (11 intermediate revisions by 2 users not shown) | |||
| Line 12: | Line 12: | ||
==Project Description== | ==Project Description== | ||
| + | |||
| + | NeedleFinder is a tool for segmentation of needles from MR scans which requires manual initialization of the tip of the needle. It has been tested extensively on MR-guided gynecologic brachytherapy data, and preliminarily on MR-guided prostate biopsy data. In this project, we aim to eliminate this reliance on manual interaction and develop a completely automatic strategy to segment the needles. We have tested a CNN approach that provides good results, even if a post processing step must be implemented in order to remove some noise and to refine the obtained segmentations. | ||
| + | |||
{| class="wikitable" | {| class="wikitable" | ||
! style="text-align: left; width:27%" | Objective | ! style="text-align: left; width:27%" | Objective | ||
| Line 19: | Line 22: | ||
| | | | ||
<!-- Objective bullet points --> | <!-- Objective bullet points --> | ||
| − | + | * Refine/clear the segmentations coming from the CNN algorithm. | |
| + | * Figure out how to transfer MRIs to a server hosting the CNN code and get back the results. | ||
| | | | ||
<!-- Approach and Plan bullet points --> | <!-- Approach and Plan bullet points --> | ||
| − | + | * Clustering and morphological filters for data cleaning. | |
| − | + | * Talk with someone from the core team to figure out how to remotely process the data. | |
| | | | ||
<!-- Progress and Next steps (fill out at the end of project week) --> | <!-- Progress and Next steps (fill out at the end of project week) --> | ||
| − | + | * A bridge between data coming by the unet and the already existing NeedleFinder module was implemented. Now we are able to use the segmentations generated by the unet to initialize the NeedleFinder algorithm. The process is completely automatic. [https://github.com/needlefinder/NunetFinder/blob/master/NunetFinder.py The code is here] | |
| + | * We had an interesting discussion with Steve about DeepInfer. | ||
|} | |} | ||
==Illustrations== | ==Illustrations== | ||
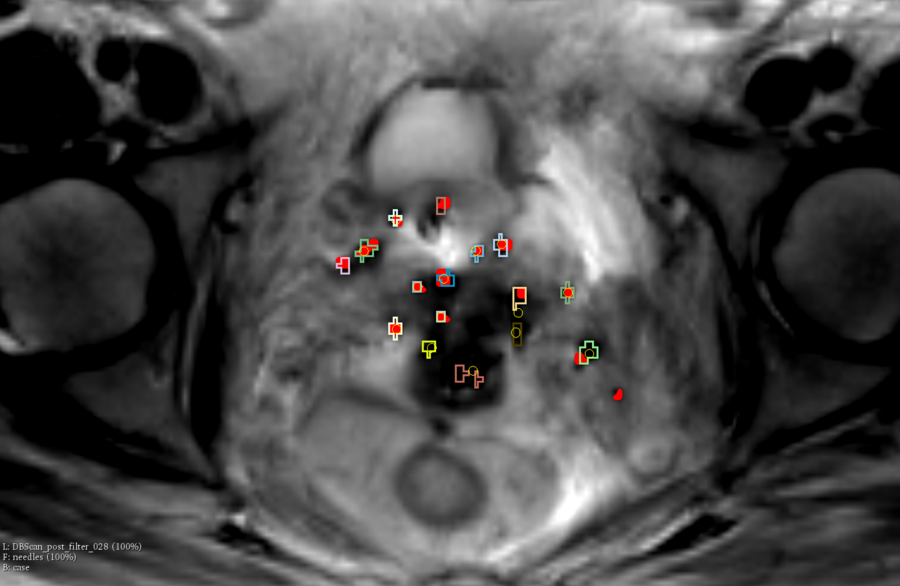
| − | + | [[File:NunetFinder.png|900px|thumb|left|Red volume: ground truth, yellow circles: needle coming from NeedleFinder, labels: segmentation coming from unet]] | |
<embedvideo service="youtube">https://youtu.be/5G9t6DZ8KrM</embedvideo> | <embedvideo service="youtube">https://youtu.be/5G9t6DZ8KrM</embedvideo> | ||
Latest revision as of 09:08, 30 June 2017
Home < Project Week 25 < NeedleSegmentation
Back to Projects List
Key Investigators
- Paolo Zaffino (Magna Graecia University, Italy)
- Salvatore Scaramuzzino (Magna Graecia University/ASL Vercelli, Italy)
- Maria Francesca Spadea (Magna Graecia University, Italy)
- Guillaume Pernelle (remote) (Imperial College, London, UK)
- Alireza Mehrtash (remote) (Brigham and Women's Hospital, Harvard Medical School, USA)
- Tina Kapur (Brigham and Women's Hospital, Harvard Medical School, USA)
Project Description
NeedleFinder is a tool for segmentation of needles from MR scans which requires manual initialization of the tip of the needle. It has been tested extensively on MR-guided gynecologic brachytherapy data, and preliminarily on MR-guided prostate biopsy data. In this project, we aim to eliminate this reliance on manual interaction and develop a completely automatic strategy to segment the needles. We have tested a CNN approach that provides good results, even if a post processing step must be implemented in order to remove some noise and to refine the obtained segmentations.
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|