Difference between revisions of "DBP2:UNC:Cortical Thickness Roadmap"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (124 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
| + | Back to [[NA-MIC_Internal_Collaborations|NA-MIC Internal Collaborations]], [[DBP2:UNC|UNC DBP 2]] | ||
| + | __NOTOC__ | ||
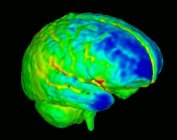
| + | [[Image:BrainDevelopment.jpg|thumb|177px|Brain development]] | ||
| + | |||
== Objective == | == Objective == | ||
We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional and local cortical thickness. | We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional and local cortical thickness. | ||
Such a workflow applied to the young brain (2-4 years old) is one goal of the [[DBP2:UNC | UNC DBP]]. This page describes the technology roadmap for cortical thickness analysis in the NA-MIC Kit. The basic components necessary for this end-to-end application are: | Such a workflow applied to the young brain (2-4 years old) is one goal of the [[DBP2:UNC | UNC DBP]]. This page describes the technology roadmap for cortical thickness analysis in the NA-MIC Kit. The basic components necessary for this end-to-end application are: | ||
| − | * Tissue segmentation: Should be multi-modality, correcting for | + | :* '''Tissue segmentation:''' Should be multi-modality, correcting for intensity inhomogeneity and work on non-skull-stripped data. |
| − | * Cortical thickness measurement: Local cortical thickness needs measurements at every location of the white-gray matter boundary, as well as at the gray-csf boundary. Regional analysis does not need such a dense measurement. | + | :* '''Cortical thickness measurement and other surface measurements (sulcal depth, surface area):''' Local cortical thickness needs measurements at every location of the white-gray matter boundary, as well as at the gray-csf boundary. Regional analysis does not need such a dense measurement. |
| − | * Cortical correspondence: Local analysis needs a full correspondence on both white-gray boundary and gray-csf boundary. | + | :* '''Cortical correspondence:''' Local analysis needs a full correspondence on both white-gray boundary and gray-csf boundary. |
| − | * Statistical analysis/Hypothesis testing: Measurements need to be compared and tested localy incorporating multiple-comparison correction, correlative analysis would be necessary too. | + | :* '''Statistical analysis/Hypothesis testing:''' Measurements need to be compared and tested localy incorporating multiple-comparison correction, correlative analysis would be necessary too. |
== Roadmap == | == Roadmap == | ||
| − | Starting with several MRI images (weighted-T1, weighted-T2...) we want to obtain cortical thickness maps for each subject, compute cortical correspondences between subjects, and analyze the cortical thickness at these corresponding locations. Ultimately, the NA-MIC Kit will provide a workflow for individual and group analysis of cortical thickness. It will be implemented as a set of Slicer3 modules that can be used interactively within the Slicer3 application as well as in batch on a computing cluster using BatchMake. | + | === Overview === |
| + | |||
| + | Starting with several MRI images (weighted-T1, weighted-T2...) we want to obtain cortical thickness maps for each subject, compute cortical correspondences between subjects, and analyze the cortical thickness at these corresponding locations. | ||
| + | Ultimately, the NA-MIC Kit will provide a workflow for individual and group analysis of cortical thickness. It will be implemented as a set of Slicer3 modules that can be used interactively within the Slicer3 application as well as in batch on a computing cluster using BatchMake. | ||
| + | |||
| + | Two pipelines have been developped: | ||
| + | :* [[DBP2:UNC:Regional_Cortical_Thickness_Pipeline | Regional cortical thickness pipeline]] | ||
| + | :**Current status: | ||
| + | :***A Slicer3 high-level module (ARCTIC) has been developed that allows individual lobar cortical thickness analysis. | ||
| + | :***ARCTIC is available to the community via NITRC. | ||
| + | :***This module has been tested on a small dataset. | ||
| + | :***Pearson correlation analysis has been performed on an adult dataset to compare ARCTIC with state of the art (FreeSurfer) | ||
| + | |||
| + | <center> | ||
| + | {| | ||
| + | |[[Image:T1Image.jpg|thumb|150px|T1-weighted skull-stripped image]] | ||
| + | |valign="center"|[[Image:Parcellation.jpg|thumb|150px|Parcellation image]] | ||
| + | |valign="center"|[[Image:WMThickness.jpg|thumb|150px|Cortical thickness on WM surface]] | ||
| + | |valign="center"|[[Image:ThicknessInformation.jpg|thumb|150px|Cortical thickness information]] | ||
| + | |} | ||
| + | </center> | ||
| + | |||
| + | :* [[DBP2:UNC:Local_Cortical_Thickness_Pipeline | Local cortical thickness pipeline]] | ||
| + | :**Current status: | ||
| + | :***A Slicer3 high-level module (GAMBIT) has been developed that allows group-wise automatic mesh-based analysis of cortical thickness. | ||
| + | :***GAMBIT is available to the community via NITRC. | ||
| + | :***This module has been tested on a small dataset. | ||
| + | |||
| + | <center> | ||
| + | {| | ||
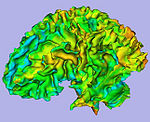
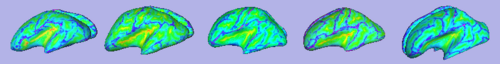
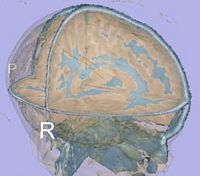
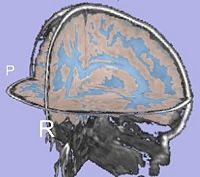
| + | |[[Image:GAMBIT QC CorticalThickness.png|thumb|500px|Cortical thickness overlayed on inflated cortical surfaces]] | ||
| + | |valign="center"|[[Image:GAMBIT_QC_SulcalDepth.png|thumb|500px|Sucal depth overlayed on inflated cortical surfaces]] | ||
| + | |} | ||
| + | </center> | ||
| + | |||
| + | |||
| + | Next we discuss the main modules and details of current status and development work: | ||
| + | |||
| + | === White matter/gray matter segmentation === | ||
| + | |||
| + | :* UNC has an open source segmentation tool called itkEMS ([http://www.ia.unc.edu/dev/download/itkems/index.htm binary download]) implemented in an ITK framework for segmenting white matter and gray matter in the young brain. An initial Slicer3 [https://www.slicer.org/wiki/Slicer3:Execution_Model_Documentation command line module] was implemented and works well. This initial Slicer3 command line module is based on the existing itkEMS xml I/O for parameters. A more extensive adaptation that would allow the direct modification of the itkEMS parameters within Slicer3 is in discussion. | ||
| + | :** Since this segmentation technique exists in an ITK framework, the integration into Slicer3 was low risk and has been completed. | ||
| + | :* UNC also adapts the current [[https://www.slicer.org/wiki/Slicer3:Execution_Model_Documentation Slicer3 EM Segment module]] to their young brain studies. Here, UNC adapted their UNC atlas of the 2 year old brain to provide priors for the EM Segment module. Initial tests have been successful, but more tests are needed. | ||
| + | :** This is a good test case for applying the Slicer3 EM Segment module to a slightly different application. UNC worked through the training material on the Slicer3 EM Segment module and discussed open issues regularly with Brad Davis (as well as Kilian Pohl). | ||
| + | :* A [[DBP2:UNC:Tissue_Segmentation_Evaluation | tissue segmentation evaluation]] was performed to compare both algorithms: itkEMS and EMSegment | ||
| − | + | <center> | |
| − | + | {| | |
| − | : | + | |[[Image:TissueSegmentation_Pediatric_EMSegment.jpg|thumb|200px|EMSegment tissue segmentation on pediatric case]] |
| − | + | |valign="top"|[[Image:TissueSegmentation_Pediatric_itkEMS.jpg|thumb|200px|itkEMS tissue segmentation on pediatric case]] | |
| − | + | |} | |
| − | + | </center> | |
| − | + | ||
| − | :* UNC has an algorithm to measure local cortical thickness given a labeling of white matter and gray matter. This technique | + | === Cortical thickness measurement === |
| − | :** This technique is | + | |
| − | :** | + | :* UNC has an algorithm to measure a local cortical thickness given a labeling of white matter and gray matter. This technique has been converted into a Slicer3 command line module. |
| − | :* Marc Niethammer | + | :** This technique is asymmetric and sparse (only computing distances where they can be computed reliably). |
| − | + | :** This module is well suited for regional analysis, but additional work to interpolate dense measurements along the boundaries from the sparse ones would be necessary for a local analysis. | |
| − | :* Regional as well as local subject comparisons are needed | + | :** A Slicer 3 external module has been implemented. |
| − | :* Regional analysis will require precise deformable registration to a young brain atlas | + | :* Marc Niethammer developed a [[Algorithm:Harvard:Thickness Slicer3 Module|technique]] at a previous project week that is symmetric. We applied his tool in first tests successfully and this module seems to offer an alternative way to compute cortical thickness measurements for a local, symmetric analysis rather than regional analysis. |
| − | :** NA-MIC | + | :** This module is well-suited for local manually-defined Regions Of Interest (ROIs). Based on linear matrices solutions, this module does not work properly when applied on the whole cortex. |
| − | :* Local analysis requires techniques which are not currently in the NA-MIC Kit | + | :** An iterative laplacian based method has been developped, as well as preprocessing modules applied on tissue segmentation label maps (largest component filter...) |
| − | :** Freesurfer | + | |
| − | :** Ipek is developing local analysis tools and | + | <center> |
| − | + | {| | |
| − | + | |[[Image:CorticalThickness_WhiteMatter.jpg|thumb|170px| Regional cortical thickness analysis on white matter surface]] | |
| − | :* | + | |valign="top"|[[Image:CorticalThickness_GreyMatter.jpg|thumb|170px|Regional cortical thickness analysis on grey matter surface]] |
| − | + | |} | |
| + | </center> | ||
| + | |||
| + | === Local correspondence === | ||
| + | :* Regional as well as local subject comparisons are needed. | ||
| + | :* Regional analysis will require precise deformable registration to a young brain atlas. | ||
| + | :** NA-MIC toolkit can be applied here, Slicer 3 has a B-spline based MI registration which needs to be tested. | ||
| + | :*** The Slicer 3 registration modules have no possibility to save/load the computed transform (only the transformed image is saved), this is a necessity for this application! | ||
| + | :*** This is an issue for both rigid, affine and b-spline based registration (all of these are needed for the full pipeline). | ||
| + | :*** Testing will be performed to see whether registration quality is appropriate | ||
| + | :*** An initial framework for the storage/retrieval and application of linear transforms has been implemented, and tests at UNC will soon take place. | ||
| + | :*** Report: Slicer3 modules (RegisterImages and ResampleScalar/Vector/DWIVolume) which allow atlas registration and image resampling are employed. | ||
| + | :** DBP has created human brain atlases (pediatric, adult, elderly) which are available to the community via MIDAS. | ||
| + | :* Local analysis requires techniques which are not currently in the NA-MIC Kit, specifically a point-to-point correspondence on the cortex (either white/gray or gray/csf) boundary needs to be established. | ||
| + | :** Freesurfer can be used for the local analysis (but it is not in the NA-MIC Kit). | ||
| + | :** Ipek Oguz is developing local analysis tools and should be available in Fall 2008. The tool necessitates the following steps (also performed within Freesurfer): | ||
| + | :*# Spherical topology of white matter: Marc Niethammer has developped a Slicer module for this step, testing will be necessary to ensure appropriateness in the cortical case. Initial tests of that module were successful. | ||
| + | :*# Inflation of cortical surface: a Slicer module exists for this step, but likely not with the right properties. DBP has implemented a Slicer3 module, with successful tests on our dataset. | ||
| + | :*# Computation of correspondence on inflated surface using particle system: No module exists, a first command line prototype tool has been developed in collaboration with Utah (I Oguz, J Cates). Initial tests have shown to be very promising (ISBI 08 submission). | ||
| − | === | + | === Hypothesis testing === |
| − | |||
| − | |||
| − | + | Regional testing can be done with standard statistical tools as there are a limited, relatively small number of regions. | |
| − | + | For local analysis, such standard statistical tools are not applicable. Currently there is no local hypothesis testing framework with multiple comparison and generalized linear model computation in Slicer3. Freesurfer has such a framework readily available. The UNC shape analysis testing framework and extensions of it should be applicable here. | |
| − | * | + | :* Freesurfer can compute group difference and correlative analysis on local cortical thickness. |
| − | * | + | :* UNC shape analysis testing framework allows local hypothesis testing with multiple comparison correction, but no General Linear Model. An extension in this regard is currently in development (M Styner). A Slicer 3 module allowing of the UNC shape analysis framework is now available. A more generic statistical testing module for Slicer 3 is in development. |
| + | :* An alternative possible testing framework is also being developed at UNC (outside of NAMIC) as an open source tool. | ||
| − | === | + | === Performance characterization and validation === |
| + | :* Characterize response based on signal noise, patient motion, etc. | ||
| + | :* Comparison to other tools (FreeSurfer, itkEMS, UNC cortical thickness). | ||
| − | + | === Schedule === | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | * Tissue segmentation | ||
| + | :* '''09/11/2007''' - Slicer registration modules input/output transforms (J Miller) -- DONE -- | ||
| + | :* '''09/11/2007''' - Slicer module to resample data through a transformation (J Miller) -- DONE -- | ||
| + | :* '''09/11/2007''' - B-spline transform nodes (J Miller) -- DONE -- | ||
| + | :* '''10/15/2007''' - White matter/gray matter segmentation of the young brain using itkEMS as a Slicer3 module (C Vachet) -- DONE -- | ||
| + | :* '''01/01/2008''' - White matter/gray matter segmentation of the young brain using the Slicer3 EM Segment module (C Vachet) -- DONE -- | ||
| + | :* '''01/02/2008''' - Separate use case scenario wiki page: how to adjust/adapt the atlas definition in the EM Segment module ([https://www.slicer.org/wiki/Modules:EM-Segmenter-Documentation:FAQ EMSegment FAQ]) (S Jaume) -- DONE -- | ||
| + | :* '''01/07/2008''' - Registration to atlas in EM Segment (B Davis) -- DONE -- | ||
| + | :* '''02/15/2008''' - [[DBP2:UNC:Tissue_Segmentation_Evaluation | Comparison study itkEMS vs EM Segment in Slicer 3]] (C Vachet) -- DONE -- | ||
| + | * Cortical thickness | ||
| + | :* '''12/23/2007''' - UNC Cortical thickness measurement as a Slicer3 module (C Vachet) -- DONE -- | ||
| + | :* '''09/01/2008''' - Niethammer's Laplacian Cortical thickness measurement module code working at UNC as a Slicer3 module (C Vachet) -- DONE -- | ||
| + | :* '''09/15/2008''' - Regional comparison between UNC and Laplacian cortical thickness (C Vachet) -- DONE -- | ||
| + | * Cortical correspondence | ||
| + | :* '''08/15/2008''' - Slicer 3 Deformable registration of young brain regional atlas (C Vachet, C Mathieu) -- DONE -- | ||
| + | :* '''11/01/2007''' - Niethammer Spherical topology tool tested on cortical data (I Oguz) -- DONE -- | ||
| + | :* '''10/01/2008''' - Inflation/Unfolding module tested on cortical data (C Vachet) -- DONE -- | ||
| + | :* '''06/01/2008''' - Slicer 3 module for cortical correspondence on inflated surface (I Oguz) -- DONE -- | ||
| + | :* '''04/01/2009''' - [[DBP2:UNC:Cortical_Thickness_Comparison | Evaluation of regional cortical thickness computation on small set of autism DBP datasets]] (C Vachet, C Mathieu) -- DONE -- | ||
| + | * Statistical analysis and Hypothesis testing | ||
| + | :* '''09/01/2008''' - GLM model for UNC statistical shape analysis (M Styner, N Augier, M Niethammer, M Macenco) -- DONE -- | ||
| + | :* '''09/01/2009''' - Slicer 3 Statistical Cortical Thickness Analysis module (C Vachet) | ||
| + | :* '''xx/xx/xxxx''' - Nonparametric hypothesis testing in ITK (J Miller) | ||
| + | * Workflow/cohesive tools | ||
| + | :* '''06/01/2008''' - Slicer 3 Workflow Shape Analysis module (C Vachet, N Augier) -- DONE --- | ||
| + | :* '''10/01/2008''' - [[DBP2:UNC:Regional_Cortical_Thickness_Pipeline | Individual regional analysis of cortical thickness as a single high-level Slicer3 module]] (C Vachet, C Mathieu) -- DONE -- | ||
| + | :* '''01/01/2010''' - [[DBP2:UNC:Local_Cortical_Thickness_Pipeline | Group-wise mesh-based analysis of cortical thickness as a NA-MIC Workflow]] (C Vachet) -- DONE -- | ||
| − | + | == Team and Institute == | |
*Co-PI: Heather Cody Hazlett, PhD, (heather_cody at med.unc.edu, Ph: 919-966-4099) | *Co-PI: Heather Cody Hazlett, PhD, (heather_cody at med.unc.edu, Ph: 919-966-4099) | ||
*Co-PI: Joseph Piven, MD | *Co-PI: Joseph Piven, MD | ||
*NA-MIC Engineering Contact: Jim Miller, GE Research | *NA-MIC Engineering Contact: Jim Miller, GE Research | ||
*NA-MIC Algorithms Contact: Martin Styner, UNC | *NA-MIC Algorithms Contact: Martin Styner, UNC | ||
| + | * Other team members | ||
| + | ** UNC DBP: Clement Vachet | ||
| + | ** UNC Algorithm: Marc Niethammer, Ipek Oguz, Nicolas Augier, Marcel Prastawa, Cedric Mathieu | ||
| + | |||
| + | |||
| + | [[Category: MRI]] [[Category: Autism]] [[Category: Segmentation]] | ||
Latest revision as of 17:00, 10 July 2017
Home < DBP2:UNC:Cortical Thickness RoadmapBack to NA-MIC Internal Collaborations, UNC DBP 2
Objective
We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional and local cortical thickness. Such a workflow applied to the young brain (2-4 years old) is one goal of the UNC DBP. This page describes the technology roadmap for cortical thickness analysis in the NA-MIC Kit. The basic components necessary for this end-to-end application are:
- Tissue segmentation: Should be multi-modality, correcting for intensity inhomogeneity and work on non-skull-stripped data.
- Cortical thickness measurement and other surface measurements (sulcal depth, surface area): Local cortical thickness needs measurements at every location of the white-gray matter boundary, as well as at the gray-csf boundary. Regional analysis does not need such a dense measurement.
- Cortical correspondence: Local analysis needs a full correspondence on both white-gray boundary and gray-csf boundary.
- Statistical analysis/Hypothesis testing: Measurements need to be compared and tested localy incorporating multiple-comparison correction, correlative analysis would be necessary too.
Roadmap
Overview
Starting with several MRI images (weighted-T1, weighted-T2...) we want to obtain cortical thickness maps for each subject, compute cortical correspondences between subjects, and analyze the cortical thickness at these corresponding locations. Ultimately, the NA-MIC Kit will provide a workflow for individual and group analysis of cortical thickness. It will be implemented as a set of Slicer3 modules that can be used interactively within the Slicer3 application as well as in batch on a computing cluster using BatchMake.
Two pipelines have been developped:
- Regional cortical thickness pipeline
- Current status:
- A Slicer3 high-level module (ARCTIC) has been developed that allows individual lobar cortical thickness analysis.
- ARCTIC is available to the community via NITRC.
- This module has been tested on a small dataset.
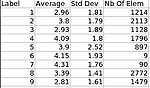
- Pearson correlation analysis has been performed on an adult dataset to compare ARCTIC with state of the art (FreeSurfer)
- Current status:
- Regional cortical thickness pipeline
- Local cortical thickness pipeline
- Current status:
- A Slicer3 high-level module (GAMBIT) has been developed that allows group-wise automatic mesh-based analysis of cortical thickness.
- GAMBIT is available to the community via NITRC.
- This module has been tested on a small dataset.
- Current status:
- Local cortical thickness pipeline
Next we discuss the main modules and details of current status and development work:
White matter/gray matter segmentation
- UNC has an open source segmentation tool called itkEMS (binary download) implemented in an ITK framework for segmenting white matter and gray matter in the young brain. An initial Slicer3 command line module was implemented and works well. This initial Slicer3 command line module is based on the existing itkEMS xml I/O for parameters. A more extensive adaptation that would allow the direct modification of the itkEMS parameters within Slicer3 is in discussion.
- Since this segmentation technique exists in an ITK framework, the integration into Slicer3 was low risk and has been completed.
- UNC also adapts the current [Slicer3 EM Segment module] to their young brain studies. Here, UNC adapted their UNC atlas of the 2 year old brain to provide priors for the EM Segment module. Initial tests have been successful, but more tests are needed.
- This is a good test case for applying the Slicer3 EM Segment module to a slightly different application. UNC worked through the training material on the Slicer3 EM Segment module and discussed open issues regularly with Brad Davis (as well as Kilian Pohl).
- A tissue segmentation evaluation was performed to compare both algorithms: itkEMS and EMSegment
- UNC has an open source segmentation tool called itkEMS (binary download) implemented in an ITK framework for segmenting white matter and gray matter in the young brain. An initial Slicer3 command line module was implemented and works well. This initial Slicer3 command line module is based on the existing itkEMS xml I/O for parameters. A more extensive adaptation that would allow the direct modification of the itkEMS parameters within Slicer3 is in discussion.
Cortical thickness measurement
- UNC has an algorithm to measure a local cortical thickness given a labeling of white matter and gray matter. This technique has been converted into a Slicer3 command line module.
- This technique is asymmetric and sparse (only computing distances where they can be computed reliably).
- This module is well suited for regional analysis, but additional work to interpolate dense measurements along the boundaries from the sparse ones would be necessary for a local analysis.
- A Slicer 3 external module has been implemented.
- Marc Niethammer developed a technique at a previous project week that is symmetric. We applied his tool in first tests successfully and this module seems to offer an alternative way to compute cortical thickness measurements for a local, symmetric analysis rather than regional analysis.
- This module is well-suited for local manually-defined Regions Of Interest (ROIs). Based on linear matrices solutions, this module does not work properly when applied on the whole cortex.
- An iterative laplacian based method has been developped, as well as preprocessing modules applied on tissue segmentation label maps (largest component filter...)
- UNC has an algorithm to measure a local cortical thickness given a labeling of white matter and gray matter. This technique has been converted into a Slicer3 command line module.
Local correspondence
- Regional as well as local subject comparisons are needed.
- Regional analysis will require precise deformable registration to a young brain atlas.
- NA-MIC toolkit can be applied here, Slicer 3 has a B-spline based MI registration which needs to be tested.
- The Slicer 3 registration modules have no possibility to save/load the computed transform (only the transformed image is saved), this is a necessity for this application!
- This is an issue for both rigid, affine and b-spline based registration (all of these are needed for the full pipeline).
- Testing will be performed to see whether registration quality is appropriate
- An initial framework for the storage/retrieval and application of linear transforms has been implemented, and tests at UNC will soon take place.
- Report: Slicer3 modules (RegisterImages and ResampleScalar/Vector/DWIVolume) which allow atlas registration and image resampling are employed.
- DBP has created human brain atlases (pediatric, adult, elderly) which are available to the community via MIDAS.
- NA-MIC toolkit can be applied here, Slicer 3 has a B-spline based MI registration which needs to be tested.
- Local analysis requires techniques which are not currently in the NA-MIC Kit, specifically a point-to-point correspondence on the cortex (either white/gray or gray/csf) boundary needs to be established.
- Freesurfer can be used for the local analysis (but it is not in the NA-MIC Kit).
- Ipek Oguz is developing local analysis tools and should be available in Fall 2008. The tool necessitates the following steps (also performed within Freesurfer):
- Spherical topology of white matter: Marc Niethammer has developped a Slicer module for this step, testing will be necessary to ensure appropriateness in the cortical case. Initial tests of that module were successful.
- Inflation of cortical surface: a Slicer module exists for this step, but likely not with the right properties. DBP has implemented a Slicer3 module, with successful tests on our dataset.
- Computation of correspondence on inflated surface using particle system: No module exists, a first command line prototype tool has been developed in collaboration with Utah (I Oguz, J Cates). Initial tests have shown to be very promising (ISBI 08 submission).
Hypothesis testing
Regional testing can be done with standard statistical tools as there are a limited, relatively small number of regions.
For local analysis, such standard statistical tools are not applicable. Currently there is no local hypothesis testing framework with multiple comparison and generalized linear model computation in Slicer3. Freesurfer has such a framework readily available. The UNC shape analysis testing framework and extensions of it should be applicable here.
- Freesurfer can compute group difference and correlative analysis on local cortical thickness.
- UNC shape analysis testing framework allows local hypothesis testing with multiple comparison correction, but no General Linear Model. An extension in this regard is currently in development (M Styner). A Slicer 3 module allowing of the UNC shape analysis framework is now available. A more generic statistical testing module for Slicer 3 is in development.
- An alternative possible testing framework is also being developed at UNC (outside of NAMIC) as an open source tool.
Performance characterization and validation
- Characterize response based on signal noise, patient motion, etc.
- Comparison to other tools (FreeSurfer, itkEMS, UNC cortical thickness).
Schedule
- Tissue segmentation
- 09/11/2007 - Slicer registration modules input/output transforms (J Miller) -- DONE --
- 09/11/2007 - Slicer module to resample data through a transformation (J Miller) -- DONE --
- 09/11/2007 - B-spline transform nodes (J Miller) -- DONE --
- 10/15/2007 - White matter/gray matter segmentation of the young brain using itkEMS as a Slicer3 module (C Vachet) -- DONE --
- 01/01/2008 - White matter/gray matter segmentation of the young brain using the Slicer3 EM Segment module (C Vachet) -- DONE --
- 01/02/2008 - Separate use case scenario wiki page: how to adjust/adapt the atlas definition in the EM Segment module (EMSegment FAQ) (S Jaume) -- DONE --
- 01/07/2008 - Registration to atlas in EM Segment (B Davis) -- DONE --
- 02/15/2008 - Comparison study itkEMS vs EM Segment in Slicer 3 (C Vachet) -- DONE --
- Cortical thickness
- 12/23/2007 - UNC Cortical thickness measurement as a Slicer3 module (C Vachet) -- DONE --
- 09/01/2008 - Niethammer's Laplacian Cortical thickness measurement module code working at UNC as a Slicer3 module (C Vachet) -- DONE --
- 09/15/2008 - Regional comparison between UNC and Laplacian cortical thickness (C Vachet) -- DONE --
- Cortical correspondence
- 08/15/2008 - Slicer 3 Deformable registration of young brain regional atlas (C Vachet, C Mathieu) -- DONE --
- 11/01/2007 - Niethammer Spherical topology tool tested on cortical data (I Oguz) -- DONE --
- 10/01/2008 - Inflation/Unfolding module tested on cortical data (C Vachet) -- DONE --
- 06/01/2008 - Slicer 3 module for cortical correspondence on inflated surface (I Oguz) -- DONE --
- 04/01/2009 - Evaluation of regional cortical thickness computation on small set of autism DBP datasets (C Vachet, C Mathieu) -- DONE --
- Statistical analysis and Hypothesis testing
- 09/01/2008 - GLM model for UNC statistical shape analysis (M Styner, N Augier, M Niethammer, M Macenco) -- DONE --
- 09/01/2009 - Slicer 3 Statistical Cortical Thickness Analysis module (C Vachet)
- xx/xx/xxxx - Nonparametric hypothesis testing in ITK (J Miller)
- Workflow/cohesive tools
- 06/01/2008 - Slicer 3 Workflow Shape Analysis module (C Vachet, N Augier) -- DONE ---
- 10/01/2008 - Individual regional analysis of cortical thickness as a single high-level Slicer3 module (C Vachet, C Mathieu) -- DONE --
- 01/01/2010 - Group-wise mesh-based analysis of cortical thickness as a NA-MIC Workflow (C Vachet) -- DONE --
Team and Institute
- Co-PI: Heather Cody Hazlett, PhD, (heather_cody at med.unc.edu, Ph: 919-966-4099)
- Co-PI: Joseph Piven, MD
- NA-MIC Engineering Contact: Jim Miller, GE Research
- NA-MIC Algorithms Contact: Martin Styner, UNC
- Other team members
- UNC DBP: Clement Vachet
- UNC Algorithm: Marc Niethammer, Ipek Oguz, Nicolas Augier, Marcel Prastawa, Cedric Mathieu