Difference between revisions of "2015 Summer Project Week:T1 mapping"
From NAMIC Wiki
Stevedaxiao (talk | contribs) |
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
||
| (12 intermediate revisions by one other user not shown) | |||
| Line 4: | Line 4: | ||
Image:T1_Mapping_logo.png|T1 Mapping Logo | Image:T1_Mapping_logo.png|T1 Mapping Logo | ||
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| Line 15: | Line 11: | ||
==Project Description== | ==Project Description== | ||
T1 mapping estimates effective tissue parameter maps (T1) from multi-spectral FLASH MRI scans with different flip angles. T1 mapping can be used to optimize parameters for a sequence, monitor diseased tissue, measure Ktrans in DCE-MRI and etc. | T1 mapping estimates effective tissue parameter maps (T1) from multi-spectral FLASH MRI scans with different flip angles. T1 mapping can be used to optimize parameters for a sequence, monitor diseased tissue, measure Ktrans in DCE-MRI and etc. | ||
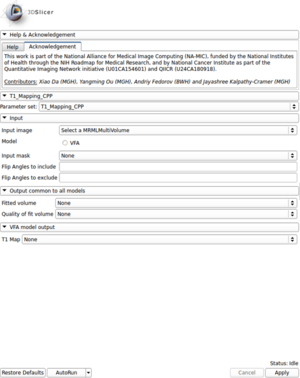
| + | [[File:T1_Mapping_CPP_GUI.png|300px|thumb|left|T1 Mapping C++ GUI]] | ||
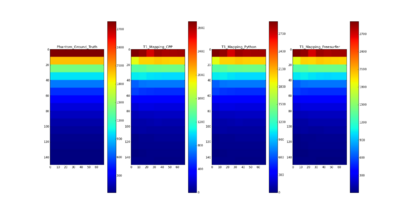
| + | [[File:Comparison_T1_Mapping_ALL.png|400px|thumb|left|Comparison of Different T1 Mapping Tools]] | ||
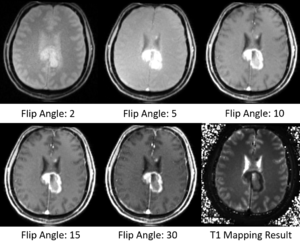
| + | [[File:T1_Mapping_Result_Sample.png|300px|thumb|left|Sample Results of T1 Mapping]] | ||
| + | |||
| + | == == | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
* Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles | * Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles | ||
| − | * Implement T1 mapping algorithm as a Slicer | + | * Implement T1 mapping algorithm as a Slicer module using C++ |
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | * Start with [ | + | * Start with [https://www.slicer.org/wiki/Documentation/Nightly/Modules/DWModeling prostate diffusion module] |
* Update equations for T1 mapping | * Update equations for T1 mapping | ||
| − | * Compare the results using C++, [ | + | * Compare the results using C++, [https://www.slicer.org/wiki/Documentation/Nightly/Modules/T1_Mapping Python], [https://surfer.nmr.mgh.harvard.edu/fswiki/mri_ms_fitparms Freesurfer ] with the ground truth of T1 for [https://dblab.duhs.duke.edu/modules/QIBAcontent/index.php?id=1 QIBA phantom] |
* Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles | * Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles | ||
| Line 31: | Line 32: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | * Updated the equations for variable flip angle (VFA) T1 mapping |
| − | * | + | * Compared the results using C++ with Python, Freesurfer and the ground truth of T1 for QIBA phantom data |
| − | * | + | * T1 mapping results on QIBA phantom data using C++ are comparable with the ground truth and the results using Freesurfer and Python |
| + | * Did some tests on MGH Brain Tumor MR Data with multiple flip angles | ||
| + | * Created a module for T1 mapping using C++ | ||
| + | ** Take multi-spectral FLASH images with an arbitrary number of flip angles as input, and estimate the T1 values of the data for each voxel | ||
| + | ** Read repetition time(TR), echo time(TE) and flip angles from the Dicom header automatically | ||
| + | ** Provide users with options to use ROI mask and choose which flip angles to include or exclude for the fitting process | ||
| + | ** Output the fitting volume and quality of fitting image as well | ||
| + | * Uploaded the source code on [https://github.com/stevedaxiao/T1_Mapping_CPP.git Github] | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 39: | Line 47: | ||
==References== | ==References== | ||
| − | * [https:// | + | * [https://sites.duke.edu/dblab/qibacontent/ QIBA T1 phantom] |
* [http://europepmc.org/articles/pmc3620726 Basic equations for T1 Mapping] | * [http://europepmc.org/articles/pmc3620726 Basic equations for T1 Mapping] | ||
* [https://github.com/stevedaxiao/T1_Mapping.git Source code for T1 Mapping Python Version] | * [https://github.com/stevedaxiao/T1_Mapping.git Source code for T1 Mapping Python Version] | ||
* [https://github.com/stevedaxiao/T1_Mapping_CPP.git Source code for T1 Mapping C++ Version] | * [https://github.com/stevedaxiao/T1_Mapping_CPP.git Source code for T1 Mapping C++ Version] | ||
Latest revision as of 17:11, 10 July 2017
Home < 2015 Summer Project Week:T1 mappingKey Investigators
- Xiao Da (MGH), Yangming Ou (MGH), Andriy Fedorov (BWH), Jayashree Kalpathy-Cramer (MGH)
- Utsav Pardasani (Observing)
Project Description
T1 mapping estimates effective tissue parameter maps (T1) from multi-spectral FLASH MRI scans with different flip angles. T1 mapping can be used to optimize parameters for a sequence, monitor diseased tissue, measure Ktrans in DCE-MRI and etc.
Objective
- Estimate effective T1 from multi-spectral FLASH MRI scans with different flip angles
- Implement T1 mapping algorithm as a Slicer module using C++
Approach, Plan
- Start with prostate diffusion module
- Update equations for T1 mapping
- Compare the results using C++, Python, Freesurfer with the ground truth of T1 for QIBA phantom
- Test the Slicer module on MGH Brain Tumor MR Data with multiple flip angles
Progress
- Updated the equations for variable flip angle (VFA) T1 mapping
- Compared the results using C++ with Python, Freesurfer and the ground truth of T1 for QIBA phantom data
- T1 mapping results on QIBA phantom data using C++ are comparable with the ground truth and the results using Freesurfer and Python
- Did some tests on MGH Brain Tumor MR Data with multiple flip angles
- Created a module for T1 mapping using C++
- Take multi-spectral FLASH images with an arbitrary number of flip angles as input, and estimate the T1 values of the data for each voxel
- Read repetition time(TR), echo time(TE) and flip angles from the Dicom header automatically
- Provide users with options to use ROI mask and choose which flip angles to include or exclude for the fitting process
- Output the fitting volume and quality of fitting image as well
- Uploaded the source code on Github