Difference between revisions of "DBP3:MGH"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (106 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | NA-MIC | + | Back to [[DBP3:Main|NA-MIC DBPs]] | [[Cores|NA-MIC Cores]] |
| + | __NOTOC__ | ||
| + | = Introduction = | ||
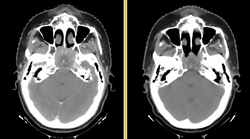
| − | + | [[File:DBP3-MGH_tumor_regression.png|250px|thumb|right|Anatomic change due to tumor regression in nasopharyngeal cancer]] | |
| − | [ | + | Head and neck cancers account for about 60,000 new cancer cases per year and represent about 6% of all cancers in the United States. These cancers are treated by a combination of chemotherapy, radiotherapy, and surgery. The five-year survival is approximately 50%. During a six-week regimen of radiotherapy, head and neck cancer patients often exhibit anatomic changes that affect their treatment. These changes include tumor regression or growth, changes in lymph node size, and changes in air cavities. Uncorrected, these changes can increase the risk of treatment complications or reduce treatment efficacy. |
| − | |||
| − | |||
| − | + | Adaptive radiotherapy is a form of personalized medicine that addresses the problem of anatomic change by incrementally adjusting the radiotherapy plan. However, a mid-treatment adjustment is complex: it requires a new CT image, image segmentation, deformable registration, and mapping of the previously delivered dose onto the new image. This project proposes to use the NA-MIC Kit to develop a simple, practical workflow for achieving personalized adaptive radiotherapy. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | = Project Goals = |
| − | |||
| − | + | This project has three specific aims: | |
| − | |||
| − | |||
| − | + | # Develop an open computational workflow for adaptive radiotherapy. | |
| − | + | # Validate the accuracy of image analysis algorithms for radiotherapy. | |
| − | + | # Evaluate the dosimetric gain of adaptive radiotherapy. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | External-beam radiotherapy requires a unique set of image analysis tools, which are only partly supported in the NA-MIC kit. Our specific aims require us to broaden the capabilities of 3D Slicer to support data interchange with commercial software, and to manipulate, review, and validate radiotherapy planning objects. We specify four sub-goals to achieve these aims. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | == DICOM-RT Interchange == |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
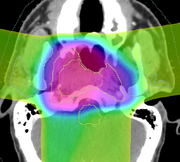
| − | + | [[File:DBP3-MGH_ss_dose.png|180px|thumb|left|RT structure set and RT dose overlayed onto CT]] | |
| − | + | DICOM-RT is an open standard exchange format for radiotherapy objects. It is well established, and is supported by nearly every modern commercial software product. The DICOM-RT data set comprises four objects: RT Dose, RT Plan, RT Structure Set, and RT Image. Briefly, RT Plan specifies the treatment beams, including beam angles, beam sizes and apertures, couch angles, and radiation output. The RT Plan is associated with a CT image through the use of the DICOM UID mechanism. An RT Dose is the output of a dose computation, measured in Gray, and is similarly associated with both a CT image and an RT Plan. The RT Structure Set contains annotations of a CT image, either points or contours, and contains the geometric boundaries of organs and structures of interest. It is associated with a CT image. Finally, the RT Image is primarily used for 2D images used in patient positioning. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Our project goal for data interchange is to provide import and export capabilities for RT Dose and RT Structure Set in the NA-MIC kit. These two DICOM items form the minimal set necessary to implement adaptive radiotherapy. Beam isocenter information is stored in the RT Plan, so we will provide RT Plan import capability as a part of plan review. RT Plan export and RT Image are outside of the scope of this project. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | === | + | == Structure and Dose Warping == |
| − | *MGH: Greg Sharp, Annie Chan, George TY Chen, Nadya Shusharina | + | |
| + | [[File:DBP3-MGH_vf_overlay.png|180px|thumb|right|Deformable registration for warping structure sets and dose]] | ||
| + | To quantify gains due to adaptive therapy, we will need to compare a plan generated on a pre-treatment CT scan with the same plans applied to a mid-treatment scan. The CT scans will be matched using deformable image registration, and registration results will be used to warp structure sets and dose. Because only the pre-treatment scan contains structure sets, plan evaluation will require mapping structures from the pre-treatment to the mid-treatment scan. Similarly, to perform dosimetric comparisons requires mapping radiation dose. | ||
| + | |||
| + | While the NA-MIC kit contains a variety of deformable image registration algorithms, the workflow for using them is not ideal for this application. Image registration in 3D Slicer creates deformed images, but we require access to the deformation maps, and a tool that can be apply previously computed deformation maps to dose and structure sets. Our goal is to improve this workflow, so that it is easy to load, save, and re-use registration results for multiple subsequent operations. Furthermore, registration algorithms that work well for one application may not work well for another, so we will benchmark and optimize existing registration algorithms for the adaptive therapy application. | ||
| + | |||
| + | == Interactive Deformable Registration == | ||
| + | |||
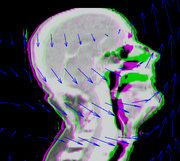
| + | [[File:DBP3-MGH_interactive_registration.png|180px|thumb|left|Interactive deformable registration with landmarks]] | ||
| + | Automatic deformable registration is the fastest and most convenient method for matching images, but there are cases where it gives sub-optimal results. In some cases, the results can be improved by adjusting the registration parameters, but in other cases the results do not improve. | ||
| + | |||
| + | To guarantee that an acceptable registration can be achieved for all cases, we propose to develop interactive registration methods using point landmarks. We will focus on point landmarks because they are conceptually simple, and they are a good match with the fiducial tools already available in 3D Slicer. The operator can control the quality of the registration by choosing the number of landmarks and adjusting their locations. | ||
| + | |||
| + | Our goal is to provide tools that can generate registration results fully automatically, fully manually, or using a combined approach. In the combined approach, we will use automatic registration as the first stage of processing, and then the operator will adjust the results using the landmark registration method. To achieve this goal, we will need an additional tool for combining the two registration results. | ||
| + | |||
| + | == Plan Review == | ||
| + | |||
| + | [[File:DBP3-MGH_dvh.png|210px|thumb|right|Plan review using dose volume histograms]] | ||
| + | Evaluation of the dosimetry results will be performed using techniques that are well established in the radiotherapy field, such as isodose evaluation and dose volume histograms (DVHs). Although it would be possible to export images from 3D Slicer back into commercial systems to perform this analysis, it is a much better workflow to do the analysis within the 3D Slicer software. The goal of this project is therefore to provide sufficient capabilities within the 3D Slicer environment, so that radiotherapy experts can perform plan review and plan comparison between a nominal treatment plan and an adaptive plan. The minimum set of useful comparison tools that we have identified to achieve this goal are isodose views, DVH generation, structure set manipulation, dose differencing, and gamma analysis. | ||
| + | |||
| + | = Research Plan = | ||
| + | |||
| + | Our research plan includes milestones for three years. | ||
| + | |||
| + | == Preliminary Work == | ||
| + | |||
| + | * Plastimatch deformable registration tool | ||
| + | ** Completed prior to start of project | ||
| + | * DICOM-RT import of RTDOSE and RTSTRUCT | ||
| + | ** Completed prior to start of project | ||
| + | |||
| + | == Year One == | ||
| + | |||
| + | * DICOM-RT export of RTDOSE and RTSTRUCT | ||
| + | ** A 3D Slicer command line executable was implemented, and is available for download from the 3D Slicer extension manager. The module is available for Windows, Linux, and OSX. DICOM-RT export has been tested for compatibility with CERR (open source) and MIM (commercial) software packages. | ||
| + | ** '''Module:''' [[2011_Winter_Project_Week:DicomRtExport|Completed 2011-01-14]]. | ||
| + | ** '''Documentation:''' (The tutorial is not yet complete.) | ||
| + | * Landmark deformable registration tool | ||
| + | ** A 3D Slicer command line executable was implemented, and is available for download from the 3D Slicer extension manager. The module is available for Windows, Linux, and OSX. The module has been tested and used for medical imaging problems in the prostate, liver and lung. | ||
| + | ** '''Module:''' [[2011_Winter_Project_Week:LandmarkRegularization|Completed 2011-01-14]]. | ||
| + | ** '''Documentation:''' [https://www.slicer.org/wiki/Modules:PlastimatchLANDWARP Completed 2011-02-14] | ||
| + | * DVH tool for plan review | ||
| + | ** '''Module:''' (The implementation is not yet complete.) | ||
| + | ** '''Documentation:''' (The tutorial is not yet complete.) | ||
| + | |||
| + | == Year Two == | ||
| + | |||
| + | * Implement V2 plastimatch deformable registration tool (loadable module -- with warping capabilities) | ||
| + | * Implement isodose overlay tool for plan review (command line executable) | ||
| + | * Implement dose differencing and gamma analysis tools for plan review (command line executable) | ||
| + | |||
| + | == Year Three == | ||
| + | |||
| + | * Implement structure set manipulation tool (loadable module) | ||
| + | * Implement V2 landmark registration tool (loadable module) | ||
| + | * Implement V2 DICOM-RT interchange tool with RTPLAN import (command line executable) | ||
| + | |||
| + | = Outreach = | ||
| + | |||
| + | == Events == | ||
| + | |||
| + | * 2011-02-11: Presentation at New England Chapter of AAPM ([http://chapter.aapm.org/NE/meet021011/talks/sharp-neaapm-2011-02-10.pdf slides]) | ||
| + | * 2011-08-02: 3D Slicer User group meeting at AAPM/COMP in Vancouver, BC | ||
| + | |||
| + | == Tutorials == | ||
| + | |||
| + | * [https://www.slicer.org/wiki/Modules:Plastimatch B-Spline deformable registration] | ||
| + | * [https://www.slicer.org/wiki/Modules:PlastimatchLANDWARP Landmark deformable registration] | ||
| + | * [https://www.slicer.org/wiki/Modules:PlastimatchDICOMRT DICOM-RT Import] | ||
| + | |||
| + | = Links = | ||
| + | |||
| + | * [http://www.na-mic.org/pages/DBP:Head_and_Neck_Cancer Project web page] | ||
| + | * [[Projects:DBP3:MGH_PlanningMeeting|Planning meeting notes]] | ||
| + | * [http://forge.abcd.harvard.edu/gf/project/plastimatch/frs/ DICOM-RT sample data] | ||
| + | * [[Algorithm:DataRT|Head and Neck Data]] | ||
| + | |||
| + | = Key Personnel = | ||
| + | *MGH: Greg Sharp, Annie Chan, George TY Chen, Nadya Shusharina | ||
*MIT: Polina Golland, Michal Depa | *MIT: Polina Golland, Michal Depa | ||
*GT: Allen Tannenbaum, Ivan Kolesov | *GT: Allen Tannenbaum, Ivan Kolesov | ||
*Isomics: Steve Pieper | *Isomics: Steve Pieper | ||
Latest revision as of 18:07, 10 July 2017
Home < DBP3:MGHBack to NA-MIC DBPs | NA-MIC Cores
Introduction
Head and neck cancers account for about 60,000 new cancer cases per year and represent about 6% of all cancers in the United States. These cancers are treated by a combination of chemotherapy, radiotherapy, and surgery. The five-year survival is approximately 50%. During a six-week regimen of radiotherapy, head and neck cancer patients often exhibit anatomic changes that affect their treatment. These changes include tumor regression or growth, changes in lymph node size, and changes in air cavities. Uncorrected, these changes can increase the risk of treatment complications or reduce treatment efficacy.
Adaptive radiotherapy is a form of personalized medicine that addresses the problem of anatomic change by incrementally adjusting the radiotherapy plan. However, a mid-treatment adjustment is complex: it requires a new CT image, image segmentation, deformable registration, and mapping of the previously delivered dose onto the new image. This project proposes to use the NA-MIC Kit to develop a simple, practical workflow for achieving personalized adaptive radiotherapy.
Project Goals
This project has three specific aims:
- Develop an open computational workflow for adaptive radiotherapy.
- Validate the accuracy of image analysis algorithms for radiotherapy.
- Evaluate the dosimetric gain of adaptive radiotherapy.
External-beam radiotherapy requires a unique set of image analysis tools, which are only partly supported in the NA-MIC kit. Our specific aims require us to broaden the capabilities of 3D Slicer to support data interchange with commercial software, and to manipulate, review, and validate radiotherapy planning objects. We specify four sub-goals to achieve these aims.
DICOM-RT Interchange
DICOM-RT is an open standard exchange format for radiotherapy objects. It is well established, and is supported by nearly every modern commercial software product. The DICOM-RT data set comprises four objects: RT Dose, RT Plan, RT Structure Set, and RT Image. Briefly, RT Plan specifies the treatment beams, including beam angles, beam sizes and apertures, couch angles, and radiation output. The RT Plan is associated with a CT image through the use of the DICOM UID mechanism. An RT Dose is the output of a dose computation, measured in Gray, and is similarly associated with both a CT image and an RT Plan. The RT Structure Set contains annotations of a CT image, either points or contours, and contains the geometric boundaries of organs and structures of interest. It is associated with a CT image. Finally, the RT Image is primarily used for 2D images used in patient positioning.
Our project goal for data interchange is to provide import and export capabilities for RT Dose and RT Structure Set in the NA-MIC kit. These two DICOM items form the minimal set necessary to implement adaptive radiotherapy. Beam isocenter information is stored in the RT Plan, so we will provide RT Plan import capability as a part of plan review. RT Plan export and RT Image are outside of the scope of this project.
Structure and Dose Warping
To quantify gains due to adaptive therapy, we will need to compare a plan generated on a pre-treatment CT scan with the same plans applied to a mid-treatment scan. The CT scans will be matched using deformable image registration, and registration results will be used to warp structure sets and dose. Because only the pre-treatment scan contains structure sets, plan evaluation will require mapping structures from the pre-treatment to the mid-treatment scan. Similarly, to perform dosimetric comparisons requires mapping radiation dose.
While the NA-MIC kit contains a variety of deformable image registration algorithms, the workflow for using them is not ideal for this application. Image registration in 3D Slicer creates deformed images, but we require access to the deformation maps, and a tool that can be apply previously computed deformation maps to dose and structure sets. Our goal is to improve this workflow, so that it is easy to load, save, and re-use registration results for multiple subsequent operations. Furthermore, registration algorithms that work well for one application may not work well for another, so we will benchmark and optimize existing registration algorithms for the adaptive therapy application.
Interactive Deformable Registration
Automatic deformable registration is the fastest and most convenient method for matching images, but there are cases where it gives sub-optimal results. In some cases, the results can be improved by adjusting the registration parameters, but in other cases the results do not improve.
To guarantee that an acceptable registration can be achieved for all cases, we propose to develop interactive registration methods using point landmarks. We will focus on point landmarks because they are conceptually simple, and they are a good match with the fiducial tools already available in 3D Slicer. The operator can control the quality of the registration by choosing the number of landmarks and adjusting their locations.
Our goal is to provide tools that can generate registration results fully automatically, fully manually, or using a combined approach. In the combined approach, we will use automatic registration as the first stage of processing, and then the operator will adjust the results using the landmark registration method. To achieve this goal, we will need an additional tool for combining the two registration results.
Plan Review
Evaluation of the dosimetry results will be performed using techniques that are well established in the radiotherapy field, such as isodose evaluation and dose volume histograms (DVHs). Although it would be possible to export images from 3D Slicer back into commercial systems to perform this analysis, it is a much better workflow to do the analysis within the 3D Slicer software. The goal of this project is therefore to provide sufficient capabilities within the 3D Slicer environment, so that radiotherapy experts can perform plan review and plan comparison between a nominal treatment plan and an adaptive plan. The minimum set of useful comparison tools that we have identified to achieve this goal are isodose views, DVH generation, structure set manipulation, dose differencing, and gamma analysis.
Research Plan
Our research plan includes milestones for three years.
Preliminary Work
- Plastimatch deformable registration tool
- Completed prior to start of project
- DICOM-RT import of RTDOSE and RTSTRUCT
- Completed prior to start of project
Year One
- DICOM-RT export of RTDOSE and RTSTRUCT
- A 3D Slicer command line executable was implemented, and is available for download from the 3D Slicer extension manager. The module is available for Windows, Linux, and OSX. DICOM-RT export has been tested for compatibility with CERR (open source) and MIM (commercial) software packages.
- Module: Completed 2011-01-14.
- Documentation: (The tutorial is not yet complete.)
- Landmark deformable registration tool
- A 3D Slicer command line executable was implemented, and is available for download from the 3D Slicer extension manager. The module is available for Windows, Linux, and OSX. The module has been tested and used for medical imaging problems in the prostate, liver and lung.
- Module: Completed 2011-01-14.
- Documentation: Completed 2011-02-14
- DVH tool for plan review
- Module: (The implementation is not yet complete.)
- Documentation: (The tutorial is not yet complete.)
Year Two
- Implement V2 plastimatch deformable registration tool (loadable module -- with warping capabilities)
- Implement isodose overlay tool for plan review (command line executable)
- Implement dose differencing and gamma analysis tools for plan review (command line executable)
Year Three
- Implement structure set manipulation tool (loadable module)
- Implement V2 landmark registration tool (loadable module)
- Implement V2 DICOM-RT interchange tool with RTPLAN import (command line executable)
Outreach
Events
- 2011-02-11: Presentation at New England Chapter of AAPM (slides)
- 2011-08-02: 3D Slicer User group meeting at AAPM/COMP in Vancouver, BC
Tutorials
Links
Key Personnel
- MGH: Greg Sharp, Annie Chan, George TY Chen, Nadya Shusharina
- MIT: Polina Golland, Michal Depa
- GT: Allen Tannenbaum, Ivan Kolesov
- Isomics: Steve Pieper