Difference between revisions of "NA-MIC Internal Collaborations:New"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (3 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
Back to [[NA-MIC_Collaborations:New|NA-MIC Collaborations]] | Back to [[NA-MIC_Collaborations:New|NA-MIC Collaborations]] | ||
__NOTOC__ | __NOTOC__ | ||
| − | = | + | = NA-MIC Internal Collaborations = |
| − | {| cellpadding="10" | + | {| cellpadding="10" border="1" |
| + | | style="width:33%" | [[Image:CingulumAllSubjectsFibers.png|300px]] | ||
| + | | style="width:33%" | [[Image:Sulcaldepth.png|300px]] | ||
| + | | style="width:33%" | [[Image:Mit_fmri_clustering_parcellation2_xsub.png|300px]] | ||
| + | |||
| + | |-align="center" | ||
| + | |||
| + | | | | ||
| + | == [[NA-MIC_Internal_Collaborations:DiffusionImageAnalysis|Diffusion Image Analysis]] == | ||
| + | |||
| + | | | | ||
| + | == [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|Structural Image Analysis]] == | ||
| + | |||
| + | | | | ||
| + | == [[NA-MIC_Internal_Collaborations:fMRIAnalysis|fMRI Analysis]] == | ||
| + | |||
| + | |} | ||
| + | |||
| + | = Driving Biological Problems (2007 - 2010): Roadmap Projects = | ||
| + | |||
| + | {| cellpadding="10" border="1" | ||
| style="width:15%" | [[Image:Cingulum1.jpg|200px]] | | style="width:15%" | [[Image:Cingulum1.jpg|200px]] | ||
| style="width:85%" | | | style="width:85%" | | ||
| Line 47: | Line 67: | ||
|} | |} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== NA-MIC Kit == | == NA-MIC Kit == | ||
=== NAMIC Software Process === | === NAMIC Software Process === | ||
| − | + | # [[NA-MIC/Projects/NA-MIC_Kit/CMake_-_NAMIC_Kit_Building|CMake - NAMIC Kit Building]] (Kitware) | |
| − | + | # [[NA-MIC/Projects/NA-MIC_Kit/CPack_-_NAMIC_Kit_Distribution|CPack - NAMIC Kit Distribution]] (Kitware) | |
| − | + | # [[NA-MIC/Projects/NA-MIC_Kit/Dart_2_and_CTest_-_Software_Quality|Dart 2 and CTest - Software Quality]] (GE, Kitware) | |
| − | | | ||
| − | |||
| − | | | ||
=== Software Infrastructure === | === Software Infrastructure === | ||
| − | + | # [[NA-MIC/Projects/NA-MIC_Kit/Slicer3|Slicer 3]] (Isomics, GE, Kitware, UCSD, UCLA) | |
| − | + | ## [[NA-MIC/Projects/NA-MIC_Kit/MRML|MRML]] (Isomics, Kitware, GE) | |
| − | | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Coordinate_Systems|Coordinate Systems]] (GE, Isomics) |
| − | | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/IO_Unification|IO Unification]] (GE, Isomics) |
| − | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Execution_Model|Execution Model]] (GE) | |
| − | | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Grid_Computing|Grid Computing]] (UCSD, GE, Kitware, Isomics) |
| + | # [[NA-MIC/Projects/NA-MIC_Kit/Licenses_Unification|Licenses Unification]] (Kitware, Isomics) | ||
| + | # Toolkits | ||
| + | ## [[NA-MIC/Projects/NA-MIC_Kit/KWWidgets|KWWidgets]] (Kitware) | ||
| + | ## [[NA-MIC/Projects/NA-MIC_Kit/Teem|Teem]] (Harvard/BWH) | ||
=== Training & Dissemination === | === Training & Dissemination === | ||
| − | + | # [[NA-MIC/Projects/NA-MIC_Kit/Training_Material_and_Workshops_for_NA-MIC_Kit|Training Material and Workshops for NA-MIC Kit]] (MGH, BWH) | |
| − | + | # [[NA-MIC/Projects/NA-MIC_Kit/Dissemination|Dissemination for NA-MIC]] (Isomics, Kitware, Harvard) | |
| − | |||
| − | |||
| − | |||
| − | | | ||
== Other Projects == | == Other Projects == | ||
| − | + | # [[Non_Rigid_Registration|Non-Rigid Registration]] | |
| − | + | # [[NA-MIC/Projects/Diffusion_Image_Analysis/FreeSurfer_NRRD_IO|FreeSurfer NRRD IO]] | |
| − | + | # [[Algorithm:MGH:FreeSurferNumericalRecipiesReplacement|FreeSurfer Numerical Recipies Replacement]] | |
| − | + | # [https://www.slicer.org/wiki/Slicer3:Fluorescence_and_Electron_Microscopy_Support Slicer3 Fluorescence and Electron Microscopy Support] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 17:08, 10 July 2017
Home < NA-MIC Internal Collaborations:NewBack to NA-MIC Collaborations
NA-MIC Internal Collaborations
|
|
|
Diffusion Image Analysis |
Structural Image Analysis |
fMRI Analysis |
Driving Biological Problems (2007 - 2010): Roadmap Projects
|
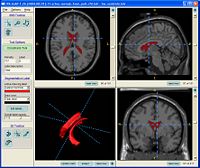
Contrasting Tractography MeasuresThis project represents a new initiative to build upon a shared vision among Cores 1, 3 and 5 that the field of medical image analysis would be well served by work in the area of validation, calibration and assessment of reliability in DW-MRI image analysis. More... New: Contrasting Tractography Methods Conference, Santa Fe, October 1-2, 2007. |
|
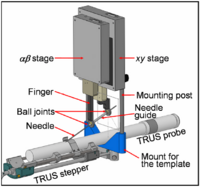
Brachytherapy Needle Positioning Robot IntegrationThe Queen’s/Hopkins team is developing novel devices and procedures for cancer interventions, including biopsy and therapies. Our goal for the programming week is to design and start implementing software for the new MRI Brachytherapy needle positioning robot. More... New: Meeting at JHU on July 17-19, 2007. |
|
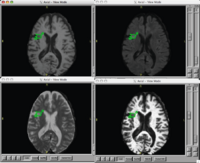
Brain Lesion Analysis in Neuropsychiatric Systemic Lupus ErythematosusOur goal is to automatically, or with little or no manual human rater input, accurately tissue classify our example lupus data-set into gray, white, csf, and lesion classes. New: Participants in the Summer 2007 NA-MIC Project Week. |
|
Cortical Thickness for AutismOur goal is to begin a longitudinal study of early brain development by cortical thickness in autistic children and controls (2 years with follow-up at 4 years). We want to be able to make statistical group comparisons. More... New: Participation in the NA-MIC Summer 2007 Project Week. |
NA-MIC Kit
NAMIC Software Process
- CMake - NAMIC Kit Building (Kitware)
- CPack - NAMIC Kit Distribution (Kitware)
- Dart 2 and CTest - Software Quality (GE, Kitware)
Software Infrastructure
- Slicer 3 (Isomics, GE, Kitware, UCSD, UCLA)
- MRML (Isomics, Kitware, GE)
- Coordinate Systems (GE, Isomics)
- IO Unification (GE, Isomics)
- Execution Model (GE)
- Grid Computing (UCSD, GE, Kitware, Isomics)
- Licenses Unification (Kitware, Isomics)
- Toolkits
Training & Dissemination
- Training Material and Workshops for NA-MIC Kit (MGH, BWH)
- Dissemination for NA-MIC (Isomics, Kitware, Harvard)