Difference between revisions of "EM Segmentation For Orthopaedic Applications"
From NAMIC Wiki
| Line 64: | Line 64: | ||
'''To Do:''' | '''To Do:''' | ||
| − | + | *Update the the tutorial to support the Slicer3 Workflow | |
| − | + | **Tutorial will be developed by Austin Ramme - In progress | |
| − | + | *Slicer3 Segmentation applied to the femur | |
| − | * Update the the tutorial to support the Slicer3 Workflow | + | **Using BRAINSFit for mapping of probability information to the subject images |
| − | ** | + | |
| − | * | + | '''Manuscript:''' |
| − | + | *Ramme AJ, Devries N, Kallemyn NA, Magnotta VA, Grosland NM. [http://www.springerlink.com/content/h972u63630311g78/ Semi-automated Phalanx Bone Segmentation Using the Expectation Maximization Algorithm]. J Digit Imaging. 2008. | |
| + | |||
'''Key Investigators:''' | '''Key Investigators:''' | ||
Latest revision as of 18:30, 20 January 2010
Home < EM Segmentation For Orthopaedic ApplicationsObjective:
- To utilize the Slicer3 Expectation Maximization Algorithm for segmentation of the phalanx bones of the hand.
Progress:
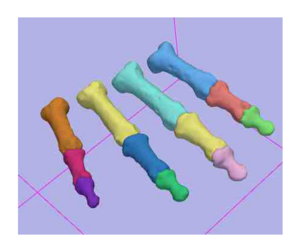
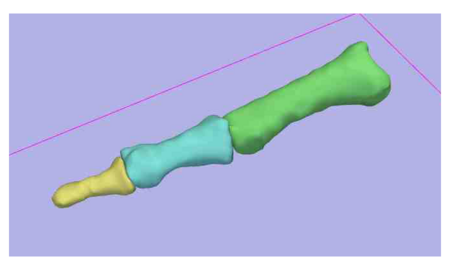
- We have utilized the Slicer2.7 EM Segmentation Module for segmentation of the phalanx bones
- Registration was performed outside of Slicer using ITK registration algorithms that are available in the IaFeMesh software. This includes Thin plate spline, B-Spline and rigid registration algorithms.
- Probability map information was created from a single subject used as the atlas image and filtered using a Gaussian filter.
- Initial evaluation has been performed
- Reliability assessed on fourteen specimens that also had the index finger segmented, and two specimens that had the index, middle, ring and little fingers manually segmented
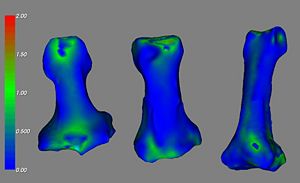
- Validation assessed using the laser scanning of the index finger (proximal, middle, and distal) phalanx bones in five specimens
- Work is underway to develop a Slicer2.7 tutorial for this segmentation (File:Draft EM Segment Tutorial 9 12.pdf)
Results:
| Finger | Phalanx Segment | Average Relative Overlap |
|---|---|---|
| Index (14 subjects) | Proximal | 0.87 |
| Medial | 0.80 | |
| Distal | 0.70 | |
| Middle (2 subjects) | Proximal | 0.79 |
| Medial | 0.77 | |
| Distal | 0.71 | |
| Ring (2 subjects) | Proximal | 0.76 |
| Medial | 0.82 | |
| Distal | 0.80 | |
| Pinky (2 subjects) | Proximal | 0.70 |
| Medial | 0.73 | |
| Distal | 0.72 |
- Example of Validation Results is shown below in the figures
To Do:
- Update the the tutorial to support the Slicer3 Workflow
- Tutorial will be developed by Austin Ramme - In progress
- Slicer3 Segmentation applied to the femur
- Using BRAINSFit for mapping of probability information to the subject images
Manuscript:
- Ramme AJ, Devries N, Kallemyn NA, Magnotta VA, Grosland NM. Semi-automated Phalanx Bone Segmentation Using the Expectation Maximization Algorithm. J Digit Imaging. 2008.
Key Investigators:
- Iowa: Austin Ramme, Nicole Grosland, and Vincent Magnotta
Links:
Figures: