Difference between revisions of "DTI Validation Telephone Conferences 2008"
RandyGollub (talk | contribs) |
RandyGollub (talk | contribs) m |
||
| (25 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | '''Tuesday April 22, 2008 - 11:00 am EST''' | ||
| + | |||
| + | '''PROGRESS:''' | ||
| + | *MIND DATA<br/> | ||
| + | The MIND data will be completely re-processed and uploaded by the end of this week. Then Sonia will verify and make any necessary corrections to the ROIs. | ||
| + | |||
| + | *PHANTOM DATA<br/> | ||
| + | The GA Tech group has converted their results to vtkPolydata format and put them into the SRB repository along with the previous nrrd format results. | ||
| + | |||
| + | The GA Tech site's Volume Tractography algorithm is undergoing another round of improvements and is starting to show good results now. Thus the currently generated results from that site should be considered preliminary and they plan to redo the analysis once the algorithm is once again in a stable state. They commented favorably on the utility of this project for testing their algorithm and giving them insights for how to make improvements. | ||
| + | |||
| + | Still waiting to for Casey, Tri and Nathan to post their TEEM phantom results on the SRB. Please just email the results to Sonia, she will get them posted and integrate them into the cross site analysis she has underway as soon as she gets them from you. | ||
| + | |||
| + | *OTHER<br/> | ||
| + | Steve Pieper briefly summarized the current status of Slicer 3 DTI processing for us. See this link for details http://www.na-mic.org/Wiki/index.php/Slicer3:DTMRI. | ||
| + | |||
| + | Carlo Pierpoli joined the meeting to hear the update on Slicer 3 and to discuss a potential future presentation on DTI analysis algorithms for the 2009 ISMRM. He will work with C.F. as the NAMIC point person to discuss further when both are at the 2008 ISMRM. | ||
| + | |||
| + | Jim Fallon sent a package with the drawn "gold standard" neural pathways we are seeking to use DTI to segment to Sonia, but we have not yet received it yet. We are working to find the package and/or he will resend. (THANKS JIM!!!) | ||
| + | |||
| + | '''ACTION PLANS''' | ||
| + | # C.-F. and John to work collaboratively to enable GA Tech to share the matlab code and TEEM libraries for running their Volumetric tractography algorithm here at BWH | ||
| + | # Next T-con is Tuesday May 13, 2008 - 11:00 am EST | ||
| + | |||
| + | |||
| + | '''Tuesday April 1, 2008 - 11:00 am EST''' | ||
| + | |||
| + | |||
| + | '''PROGRESS:''' | ||
| + | *MIND DATA<br/> | ||
| + | We are still waiting for Jeremy/MIND team to redo the whole cohort and upload the data, replacing the incorrect files on the SRB. | ||
| + | |||
| + | *PHANTOM DATA<br/> | ||
| + | Two weeks ago Casey, Tri and Nathan (in an email to Randy) reported successfully generating tracts from the TEEM phantom but they have not yet posted them on the SRB. Please just email the results to Sonia, she will get them posted and integrate them into the cross site analysis she has underway as soon as she gets them from you. John also obtained results, but isn't happy with results- the algorithm "hugs the inner curvature" of the helix. He is working with a new phantom that he made that is more suited for his algorithm. Discussed value of sharing vtkPolydata results from TEEM phantom, John will show Vandana how to do this. | ||
| + | |||
| + | *OTHER<br/> | ||
| + | CF briefly summarized results of Core 1 meeting in SLC March 24-26. See this link for details http://www.na-mic.org/Wiki/index.php/2008_Engineering_review_at_Utah:Summary. | ||
| + | |||
| + | Proceeding with plans for Randy, CF and Sonia to co-mentor a MIT summer student to assist with data analysis, project integration and presentation of results. | ||
| + | |||
| + | '''ACTION PLANS''' | ||
| + | # Process TEEM phantom data and email Sonia your results in vtkPolydata format | ||
| + | # Email your scripts that you use to process the TEEM phantom to CF so he can use them as "use cases" for Slicer3 DTI workflow tool development. | ||
| + | # Don't work on the MIND data until we notify you that the correct data is posted and ready for download. | ||
| + | # Next T-con is Tuesday April 22, 2008 - 11:00 am EST; NOTE PUSHED BACK ONE WEEK | ||
| + | |||
| + | |||
| + | '''Tuesday March 18th, 2008 - 11:00 am EST''' | ||
| + | |||
| + | |||
| + | '''PROGRESS:''' | ||
| + | *MIND DATA<br/> | ||
| + | Sonia has fixed the scripts that convert the Siemens DICOM image data to nrrd format. She is passing the scripts to Jeremy who will redo the whole cohort and upload the data, replacing the incorrect files on the SRB by the end of this week. | ||
| + | |||
| + | *PHANTOM DATA<br/> | ||
| + | Casey, Tri and Nathan (in an email to Randy) reported successfully generating tracts from the TEEM phantom and are working to post them on the SRB. See email from Jeremy regarding changes due to the SRB upgrades that have recently taken place. | ||
| + | |||
| + | *OTHER<br/> | ||
| + | Discussed integration of tractography algorithms into the Slicer 3 environment as a way to facilitate forward progress. Ross, Guido, CF, Casey, Steve, and others will be meeting in SLC March 24-26 and this will be one of the agenda items. We are all eager to see this happen. | ||
| + | |||
| + | Suggestion to consolidate analysis of data from multiple algorithms (in or out of Slicer3) at a single site (BWH hosting) considered by the group. Randy offered to sponsor from Training Core funds a summer student to assist with data analysis, project integration and presentation of results. | ||
| + | |||
| + | '''ACTION PLANS''' | ||
| + | #All groups need to finish processing the TEEM helix data and post tracts file in VTKPolydata format on SRB by next T-con (sound familiar?). Next T-con is April Fool's Day. | ||
| + | # Don't work on the MIND data until we notify you that the correct data is posted and ready for download. | ||
| + | |||
| + | |||
| + | |||
| + | '''Tuesday March 4, 2008 - 11:00 am EST''' | ||
| + | |||
| + | |||
| + | '''PROGRESS:''' | ||
| + | *MIND DATA<br/> | ||
| + | Sonia has identified that the initial script run in New Mexico to start the conversion of the DWI DICOM mosaic data to nrrd format erroneously cut 4 slices of the image data away. This is what resulted in the misregistration with the Freesurfer segmentation data and the created the problem with the acruate fasciculus ROIs. This good to know before we get any further so we can verify that this error did not affect the brain and report back next meeting. She will need to redo the ROIs so don't work on this part of the project yet. Jim Fallon is creating the "master neuroanatomist" hand segmentation of the tracts we are using for this project. (Awesome JIM!) | ||
| + | *PHANTOM DATA<br/> | ||
| + | Sonia posted Slicer tractography results from the TEEM phantom (see figure below). John M downloaded and began processing the phantom data, but is noting some issues due to the large homogenous volume of the synthetic tract (e.g. follows inner curve of helix). He would like to work with it more and will report results at next T-con. Nothing from any other site. MICCAI and other deadlines have been a major time constraint. Discussed options for how to motivate forward progress on project, with CF suggesting more frequent email reminders and use of the wiki to document individual site progress. | ||
| + | *OTHER<br/> | ||
| + | Randy described another approach to validating DTI tractography used by Martinos colleagues. She will circulate the 2005 ISMRM abstract and associated website information to the group for future discussion. | ||
| + | <br/> | ||
| + | '''ACTION PLANS''' | ||
| + | #All groups need to process the TEEM helix data and post results by next T-con on March 18th | ||
| + | #Sonia will set up a wiki progress page for us | ||
| + | #Randy will send first reminder to group right after MICCAI deadline | ||
| + | |||
| + | |||
| + | |||
| + | '''Tuesday February 19, 2008 - 11:00 am EST''' | ||
| + | |||
| + | |||
| + | '''PROGRESS:''' | ||
| + | The TEEM phantom data was downloaded by a few groups who were able to generate tracts from it. No problems were encountered, but not all groups have yet attempted it. I believe that all sites have now uploaded results for the one subject we agreed to do for the AHM meeting. Sylvain Gouttard found an error in the ROIs posted for generating the arcuate fasiculus tract, so please disregard that until Sonia posts a corrected set of ROIs and sends out an email notice. Nathan was not on call and no input on what the problems are with his phantom. | ||
| + | |||
| + | == '''ACTION PLANS''': == | ||
| + | #All sites PLEASE download new TEEM phantom data (in nrrd format w/ROIs) ASAP and run your tractography algorithm on it, and upload your results (tract fields and matricies) before next T-con (Tuesday March 4th). | ||
| + | # All sites look at the results of the registration between the DTI data and structural scan for the exemplar subject (and as many as you have time for) so we can discuss any outstanding registration issues (brought up as a potential problem by Guido). | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br/> | ||
| + | |||
| + | '''Tuesday February 5, 2008 - 11:00 am EST''' | ||
| + | |||
| + | |||
| + | '''PROGRESS:''' | ||
| + | <br/> | ||
| + | '''DWI PHANTOM'''<br/> | ||
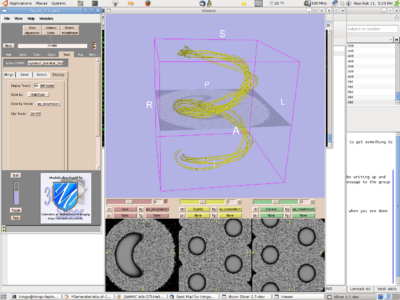
| + | [[Image:TEEMphantom.png | 400 px |right ]] | ||
| + | * Many sites were able to download Nathan's phantom data, but none could generate reasonable tractography results. | ||
| + | * Nathan was unable to join our T-con due to illness but has since been appraised of the situation and is in agreement with the group plan. | ||
| + | * Tri volunteered and has completed generating another phantom using the TEEM software. The group agreed that he would use a simple unity measurement frame, isotropic voxels, match the MIND data where possible, add "some" noise. | ||
| + | * Tri has already uploaded the phantom data (dwi and dti images and a "white matter mask") and given our group permission to view it on our SRB space in a directory called "home/Projects/NAMIC_DTI_VALIDITY__0074/Data/helix_phantom_NAMIC_DTI_validity" | ||
| + | * Sonia has generated and posted ROIs for the group to use for generating tractography results in the directory 'home/Projects/NAMIC_DTI_VALIDITY__0074/Data/helix_phantom_NAMIC_DTI_validity'. | ||
| + | <br/> | ||
| + | '''MIND DATA ANALYSIS''' | ||
| + | * Marek, Randy and Sonia met with Jim Fallon on Friday Feb.1st. With Jim's help we developed Freesurfer based methods to make cortical gray matter target ROIs for our tracts. The group agreed that we would start with a single tract, the arcuate fasiculus to test our outcome measures. The "source" is the inferior frontal gyrus and adjacent sulcus, the "sink" is the superior temporal gyrus and adjacent sulcus as delineated by the original Freesurfer cortical parcellation scheme. See Automatically Parcellating the Human Cerebral Cortex, Fischl, B., A. van der Kouwe, C. Destrieux, E. Halgren, F. Segonne, D. Salat, E. Busa, L. Seidman, J. Goldstein, D. Kennedy, V. Caviness, N. Makris, B. Rosen, and A.M. Dale, (2004). Cerebral Cortex, 14:11-22. | ||
| + | * We agreed to generate these two ROIs for the first sample subject and to "grow them" out into the underlying white matter so that algorithms such as stochastic tractography that need white matter targets can use the same ROIs. | ||
| + | * Randy and Anastasia generated the initial ROIs as requested in Freesurfer and Sonia has reformatted them for the group and posted them on the SRB in the directory 'home/Projects/NAMIC_DTI_VALIDITY__0074/Data'. | ||
| + | |||
| + | * Randy will update the wiki to add this info to our ROI page. | ||
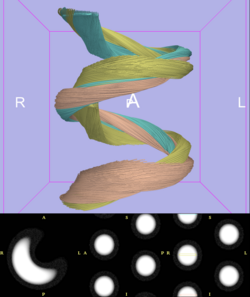
| + | [[Image:TeemPhantom-SlicerROI0001.png | 250 px | right]] | ||
| + | |||
| + | == '''ACTION PLANS''': == | ||
| + | #All sites download Tri's new phantom data (in nrrd format w/ROIs) ASAP and run your tractography algorithm on it, and upload your results (tract fields and matricies) before next Tuesday's T-con (Tuesday Feb. 19th). | ||
| + | # Nathan will work with the group to determine what problems caused tractography with his phantom to fail. (Nathan can you please post an image of a tract you generated from the phantom using your algorithm so that everyone knows what the target outcome is?) | ||
| + | # TWO SITES STILL NEED TO UPLOAD TRACTOGRAPHY RESULTS FROM VISIT 1 OF THE SAMPLE MIND DATA- YOU KNOW WHO YOU ARE!!!! | ||
| + | <br/> | ||
| + | |||
'''Tuesday January 22, 2008 - 11:00 am EST''' | '''Tuesday January 22, 2008 - 11:00 am EST''' | ||
| − | * Nathan | + | |
| + | '''PROGRESS:''' | ||
| + | * Nathan has posted both a 6 gradient direction phantom and a helical phantom that simulates a 60 gradient DWI acquisition mirrors the MIND Reliability dataset. Both are now available on the BIRN portal along with a document with a description of the different files present and how to use them is in the phantom directories: | ||
/home/Projects/NAMIC_DTI_VALIDITY__0074/Data/helix_phantom_NAMIC_DTI_validity/ | /home/Projects/NAMIC_DTI_VALIDITY__0074/Data/helix_phantom_NAMIC_DTI_validity/ | ||
| − | |||
| − | + | Both are in analyze format. Nathan is working to transform them into nrrd format, '''any help would be gratefully appreciated'''. | |
| + | * Once they are in nrrd format, Sonia will generate the Regions of Interest and post those ASAP. | ||
| − | * | + | * Nathan is creating a wiki page with complete [http://www.na-mic.org/Wiki/index.php/Hageman:NAMICHelixPhantom description of the phantom] methods and link to download the binary. |
| − | + | ||
| + | |||
| + | * Anastasia has uploaded the transformed masks and segmentations for the MIND Reliability subjects into the Data directory: | ||
| + | freesurfer_segmentations_whitemattermasks_xformed_to_corrected_dwi_visit1_all_subjects.tar.gz | ||
| + | |||
| + | and the transformation matrices: | ||
| + | freesurfer_to_corrected_dwi_visit1_xform_matrices_all_subjects.tar.gz | ||
| + | |||
| + | One could now start with the original segmentations, which are under freesurfer_results, and apply these matrices to resample to the corrected DWI visit 1 data. | ||
* Jim Fallon has has accepted an invitation to work with the Clinical team (Randy and Marek) to define and generate the additional ROIs and neuroanatomical landmarks for the tracts that we agreed upon at the AHM. They will be working together on Friday Feb 1 in Boston and will report progress on next T-con. | * Jim Fallon has has accepted an invitation to work with the Clinical team (Randy and Marek) to define and generate the additional ROIs and neuroanatomical landmarks for the tracts that we agreed upon at the AHM. They will be working together on Friday Feb 1 in Boston and will report progress on next T-con. | ||
| Line 16: | Line 154: | ||
== '''ACTION PLAN''': == | == '''ACTION PLAN''': == | ||
| − | All sites download the new phantom data and run your tractography algorithm on it before next Tuesday's T-con. | + | All sites download the new phantom data (in nrrd format w/ROIs) on Monday and run your tractography algorithm on it before next Tuesday's T-con. |
| − | Use this list to work out any logistical problems. | + | Use this email list to work out any logistical problems. |
| + | |||
| + | |||
Back to [[Projects/Diffusion/2007_Project_Week_Contrasting_Tractography_Measures | Contrasting Tractography Project Page]] | Back to [[Projects/Diffusion/2007_Project_Week_Contrasting_Tractography_Measures | Contrasting Tractography Project Page]] | ||
Latest revision as of 16:06, 22 April 2008
Home < DTI Validation Telephone Conferences 2008Tuesday April 22, 2008 - 11:00 am EST
PROGRESS:
- MIND DATA
The MIND data will be completely re-processed and uploaded by the end of this week. Then Sonia will verify and make any necessary corrections to the ROIs.
- PHANTOM DATA
The GA Tech group has converted their results to vtkPolydata format and put them into the SRB repository along with the previous nrrd format results.
The GA Tech site's Volume Tractography algorithm is undergoing another round of improvements and is starting to show good results now. Thus the currently generated results from that site should be considered preliminary and they plan to redo the analysis once the algorithm is once again in a stable state. They commented favorably on the utility of this project for testing their algorithm and giving them insights for how to make improvements.
Still waiting to for Casey, Tri and Nathan to post their TEEM phantom results on the SRB. Please just email the results to Sonia, she will get them posted and integrate them into the cross site analysis she has underway as soon as she gets them from you.
- OTHER
Steve Pieper briefly summarized the current status of Slicer 3 DTI processing for us. See this link for details http://www.na-mic.org/Wiki/index.php/Slicer3:DTMRI.
Carlo Pierpoli joined the meeting to hear the update on Slicer 3 and to discuss a potential future presentation on DTI analysis algorithms for the 2009 ISMRM. He will work with C.F. as the NAMIC point person to discuss further when both are at the 2008 ISMRM.
Jim Fallon sent a package with the drawn "gold standard" neural pathways we are seeking to use DTI to segment to Sonia, but we have not yet received it yet. We are working to find the package and/or he will resend. (THANKS JIM!!!)
ACTION PLANS
- C.-F. and John to work collaboratively to enable GA Tech to share the matlab code and TEEM libraries for running their Volumetric tractography algorithm here at BWH
- Next T-con is Tuesday May 13, 2008 - 11:00 am EST
Tuesday April 1, 2008 - 11:00 am EST
PROGRESS:
- MIND DATA
We are still waiting for Jeremy/MIND team to redo the whole cohort and upload the data, replacing the incorrect files on the SRB.
- PHANTOM DATA
Two weeks ago Casey, Tri and Nathan (in an email to Randy) reported successfully generating tracts from the TEEM phantom but they have not yet posted them on the SRB. Please just email the results to Sonia, she will get them posted and integrate them into the cross site analysis she has underway as soon as she gets them from you. John also obtained results, but isn't happy with results- the algorithm "hugs the inner curvature" of the helix. He is working with a new phantom that he made that is more suited for his algorithm. Discussed value of sharing vtkPolydata results from TEEM phantom, John will show Vandana how to do this.
- OTHER
CF briefly summarized results of Core 1 meeting in SLC March 24-26. See this link for details http://www.na-mic.org/Wiki/index.php/2008_Engineering_review_at_Utah:Summary.
Proceeding with plans for Randy, CF and Sonia to co-mentor a MIT summer student to assist with data analysis, project integration and presentation of results.
ACTION PLANS
- Process TEEM phantom data and email Sonia your results in vtkPolydata format
- Email your scripts that you use to process the TEEM phantom to CF so he can use them as "use cases" for Slicer3 DTI workflow tool development.
- Don't work on the MIND data until we notify you that the correct data is posted and ready for download.
- Next T-con is Tuesday April 22, 2008 - 11:00 am EST; NOTE PUSHED BACK ONE WEEK
Tuesday March 18th, 2008 - 11:00 am EST
PROGRESS:
- MIND DATA
Sonia has fixed the scripts that convert the Siemens DICOM image data to nrrd format. She is passing the scripts to Jeremy who will redo the whole cohort and upload the data, replacing the incorrect files on the SRB by the end of this week.
- PHANTOM DATA
Casey, Tri and Nathan (in an email to Randy) reported successfully generating tracts from the TEEM phantom and are working to post them on the SRB. See email from Jeremy regarding changes due to the SRB upgrades that have recently taken place.
- OTHER
Discussed integration of tractography algorithms into the Slicer 3 environment as a way to facilitate forward progress. Ross, Guido, CF, Casey, Steve, and others will be meeting in SLC March 24-26 and this will be one of the agenda items. We are all eager to see this happen.
Suggestion to consolidate analysis of data from multiple algorithms (in or out of Slicer3) at a single site (BWH hosting) considered by the group. Randy offered to sponsor from Training Core funds a summer student to assist with data analysis, project integration and presentation of results.
ACTION PLANS
- All groups need to finish processing the TEEM helix data and post tracts file in VTKPolydata format on SRB by next T-con (sound familiar?). Next T-con is April Fool's Day.
- Don't work on the MIND data until we notify you that the correct data is posted and ready for download.
Tuesday March 4, 2008 - 11:00 am EST
PROGRESS:
- MIND DATA
Sonia has identified that the initial script run in New Mexico to start the conversion of the DWI DICOM mosaic data to nrrd format erroneously cut 4 slices of the image data away. This is what resulted in the misregistration with the Freesurfer segmentation data and the created the problem with the acruate fasciculus ROIs. This good to know before we get any further so we can verify that this error did not affect the brain and report back next meeting. She will need to redo the ROIs so don't work on this part of the project yet. Jim Fallon is creating the "master neuroanatomist" hand segmentation of the tracts we are using for this project. (Awesome JIM!)
- PHANTOM DATA
Sonia posted Slicer tractography results from the TEEM phantom (see figure below). John M downloaded and began processing the phantom data, but is noting some issues due to the large homogenous volume of the synthetic tract (e.g. follows inner curve of helix). He would like to work with it more and will report results at next T-con. Nothing from any other site. MICCAI and other deadlines have been a major time constraint. Discussed options for how to motivate forward progress on project, with CF suggesting more frequent email reminders and use of the wiki to document individual site progress.
- OTHER
Randy described another approach to validating DTI tractography used by Martinos colleagues. She will circulate the 2005 ISMRM abstract and associated website information to the group for future discussion.
ACTION PLANS
- All groups need to process the TEEM helix data and post results by next T-con on March 18th
- Sonia will set up a wiki progress page for us
- Randy will send first reminder to group right after MICCAI deadline
Tuesday February 19, 2008 - 11:00 am EST
PROGRESS:
The TEEM phantom data was downloaded by a few groups who were able to generate tracts from it. No problems were encountered, but not all groups have yet attempted it. I believe that all sites have now uploaded results for the one subject we agreed to do for the AHM meeting. Sylvain Gouttard found an error in the ROIs posted for generating the arcuate fasiculus tract, so please disregard that until Sonia posts a corrected set of ROIs and sends out an email notice. Nathan was not on call and no input on what the problems are with his phantom.
ACTION PLANS:
- All sites PLEASE download new TEEM phantom data (in nrrd format w/ROIs) ASAP and run your tractography algorithm on it, and upload your results (tract fields and matricies) before next T-con (Tuesday March 4th).
- All sites look at the results of the registration between the DTI data and structural scan for the exemplar subject (and as many as you have time for) so we can discuss any outstanding registration issues (brought up as a potential problem by Guido).
Tuesday February 5, 2008 - 11:00 am EST
PROGRESS:
DWI PHANTOM
- Many sites were able to download Nathan's phantom data, but none could generate reasonable tractography results.
- Nathan was unable to join our T-con due to illness but has since been appraised of the situation and is in agreement with the group plan.
- Tri volunteered and has completed generating another phantom using the TEEM software. The group agreed that he would use a simple unity measurement frame, isotropic voxels, match the MIND data where possible, add "some" noise.
- Tri has already uploaded the phantom data (dwi and dti images and a "white matter mask") and given our group permission to view it on our SRB space in a directory called "home/Projects/NAMIC_DTI_VALIDITY__0074/Data/helix_phantom_NAMIC_DTI_validity"
- Sonia has generated and posted ROIs for the group to use for generating tractography results in the directory 'home/Projects/NAMIC_DTI_VALIDITY__0074/Data/helix_phantom_NAMIC_DTI_validity'.
MIND DATA ANALYSIS
- Marek, Randy and Sonia met with Jim Fallon on Friday Feb.1st. With Jim's help we developed Freesurfer based methods to make cortical gray matter target ROIs for our tracts. The group agreed that we would start with a single tract, the arcuate fasiculus to test our outcome measures. The "source" is the inferior frontal gyrus and adjacent sulcus, the "sink" is the superior temporal gyrus and adjacent sulcus as delineated by the original Freesurfer cortical parcellation scheme. See Automatically Parcellating the Human Cerebral Cortex, Fischl, B., A. van der Kouwe, C. Destrieux, E. Halgren, F. Segonne, D. Salat, E. Busa, L. Seidman, J. Goldstein, D. Kennedy, V. Caviness, N. Makris, B. Rosen, and A.M. Dale, (2004). Cerebral Cortex, 14:11-22.
- We agreed to generate these two ROIs for the first sample subject and to "grow them" out into the underlying white matter so that algorithms such as stochastic tractography that need white matter targets can use the same ROIs.
- Randy and Anastasia generated the initial ROIs as requested in Freesurfer and Sonia has reformatted them for the group and posted them on the SRB in the directory 'home/Projects/NAMIC_DTI_VALIDITY__0074/Data'.
- Randy will update the wiki to add this info to our ROI page.
ACTION PLANS:
- All sites download Tri's new phantom data (in nrrd format w/ROIs) ASAP and run your tractography algorithm on it, and upload your results (tract fields and matricies) before next Tuesday's T-con (Tuesday Feb. 19th).
- Nathan will work with the group to determine what problems caused tractography with his phantom to fail. (Nathan can you please post an image of a tract you generated from the phantom using your algorithm so that everyone knows what the target outcome is?)
- TWO SITES STILL NEED TO UPLOAD TRACTOGRAPHY RESULTS FROM VISIT 1 OF THE SAMPLE MIND DATA- YOU KNOW WHO YOU ARE!!!!
Tuesday January 22, 2008 - 11:00 am EST
PROGRESS:
- Nathan has posted both a 6 gradient direction phantom and a helical phantom that simulates a 60 gradient DWI acquisition mirrors the MIND Reliability dataset. Both are now available on the BIRN portal along with a document with a description of the different files present and how to use them is in the phantom directories:
/home/Projects/NAMIC_DTI_VALIDITY__0074/Data/helix_phantom_NAMIC_DTI_validity/
Both are in analyze format. Nathan is working to transform them into nrrd format, any help would be gratefully appreciated.
- Once they are in nrrd format, Sonia will generate the Regions of Interest and post those ASAP.
- Nathan is creating a wiki page with complete description of the phantom methods and link to download the binary.
- Anastasia has uploaded the transformed masks and segmentations for the MIND Reliability subjects into the Data directory:
freesurfer_segmentations_whitemattermasks_xformed_to_corrected_dwi_visit1_all_subjects.tar.gz
and the transformation matrices: freesurfer_to_corrected_dwi_visit1_xform_matrices_all_subjects.tar.gz
One could now start with the original segmentations, which are under freesurfer_results, and apply these matrices to resample to the corrected DWI visit 1 data.
- Jim Fallon has has accepted an invitation to work with the Clinical team (Randy and Marek) to define and generate the additional ROIs and neuroanatomical landmarks for the tracts that we agreed upon at the AHM. They will be working together on Friday Feb 1 in Boston and will report progress on next T-con.
ACTION PLAN:
All sites download the new phantom data (in nrrd format w/ROIs) on Monday and run your tractography algorithm on it before next Tuesday's T-con. Use this email list to work out any logistical problems.