Difference between revisions of "BRAINSCut"
From NAMIC Wiki
| (One intermediate revision by the same user not shown) | |||
| Line 2: | Line 2: | ||
| − | + | ===Summary=== | |
BRAINSCut is a software package for segmentation of structures using artificial neural networks. Currently this tool supports the segmentation of the following structures: brain, caudate, putamen, thalamus, hippocampus, anterior cerebellum, interior posterior cerebellum, superior posterior cerebellum, corpus medullary. Future regions will include the globus pallidus, amygdala, and nucleus accumbens. The command line uses the Slicer3 execution model framework. | BRAINSCut is a software package for segmentation of structures using artificial neural networks. Currently this tool supports the segmentation of the following structures: brain, caudate, putamen, thalamus, hippocampus, anterior cerebellum, interior posterior cerebellum, superior posterior cerebellum, corpus medullary. Future regions will include the globus pallidus, amygdala, and nucleus accumbens. The command line uses the Slicer3 execution model framework. | ||
| Line 8: | Line 8: | ||
| − | + | ===Progress=== | |
#Integration with a high dimensional registration to the atlas probability map | #Integration with a high dimensional registration to the atlas probability map | ||
#Improved thresholding of the output activation maps | #Improved thresholding of the output activation maps | ||
| Line 15: | Line 15: | ||
| − | + | ===To Do=== | |
#Complete integration with the FANN library | #Complete integration with the FANN library | ||
#Link to a BSD style neural network library | #Link to a BSD style neural network library | ||
| Line 21: | Line 21: | ||
| − | + | ===Key Investigators=== | |
* University of Iowa: Hans Johnson, Ronald Pierson, Kent Williams, Greg Harris, Vincent Magnotta | * University of Iowa: Hans Johnson, Ronald Pierson, Kent Williams, Greg Harris, Vincent Magnotta | ||
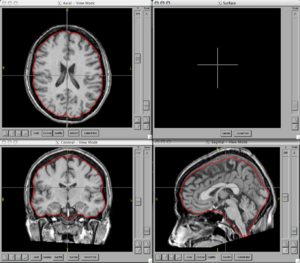
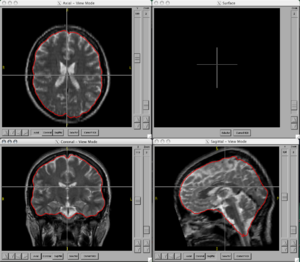
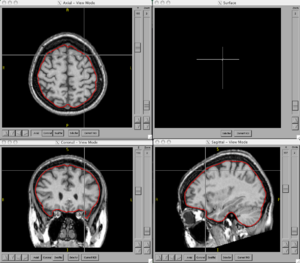
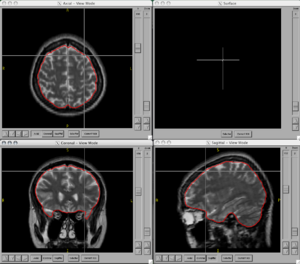
| − | + | ===Figures=== | |
| − | <gallery Caption=" | + | <gallery Caption="BRAINSCut" widths="300px" heights="300px" perrow="2"> |
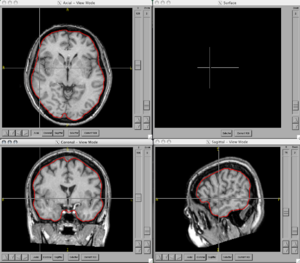
Image:BRAINS-ANN-Brain-Subject1-T1.png|Brain Mask generated using BRAINSMush image and BRAINSCut overlaid on T1 for Subject 1 | Image:BRAINS-ANN-Brain-Subject1-T1.png|Brain Mask generated using BRAINSMush image and BRAINSCut overlaid on T1 for Subject 1 | ||
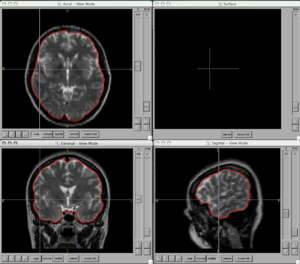
Image:BRAINS-ANN-Brain-Subject1-T2.png|Brain Mask generated using BRAINSMush image and BRAINSCut overlaid on T2 for Subject 1 | Image:BRAINS-ANN-Brain-Subject1-T2.png|Brain Mask generated using BRAINSMush image and BRAINSCut overlaid on T2 for Subject 1 | ||
| Line 37: | Line 37: | ||
</gallery> | </gallery> | ||
| − | + | ===Usage=== | |
| − | |||
| − | |||
BRAINSCut [--processinformationaddress <std::string>] [--xml] [--echo] | BRAINSCut [--processinformationaddress <std::string>] [--xml] [--echo] | ||
[--applyModel] [--trainModel] [--createVectors] | [--applyModel] [--trainModel] [--createVectors] | ||
| Line 90: | Line 88: | ||
| − | + | ===Links=== | |
*[http://www.psychiatry.uiowa.edu University of Iowa Department of Psychiatry] | *[http://www.psychiatry.uiowa.edu University of Iowa Department of Psychiatry] | ||
*[http://mri.radiology.uiowa.edu University of Iowa MRI Center] | *[http://mri.radiology.uiowa.edu University of Iowa MRI Center] | ||
| − | + | ===Papers=== | |
*Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WNP-4PGGP30-3&_user=440026&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000020939&_version=1&_urlVersion=0&_userid=440026&md5=c783cac6caa49b7253ab0d08c9da068b Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures]. Neuroimage. 39(1):238-47, 2008. | *Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WNP-4PGGP30-3&_user=440026&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000020939&_version=1&_urlVersion=0&_userid=440026&md5=c783cac6caa49b7253ab0d08c9da068b Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures]. Neuroimage. 39(1):238-47, 2008. | ||
*Magnotta VA, Heckel D, Andreasen NC, Cizadlo T, Corson PW, Ehrhardt JC, Yuh WT. [http://radiology.rsnajnls.org/cgi/content/full/211/3/781 Measurement of brain structures with artificial neural networks: two- and three-dimensional applications]. Radiology. 211(3):781-90, 1999. | *Magnotta VA, Heckel D, Andreasen NC, Cizadlo T, Corson PW, Ehrhardt JC, Yuh WT. [http://radiology.rsnajnls.org/cgi/content/full/211/3/781 Measurement of brain structures with artificial neural networks: two- and three-dimensional applications]. Radiology. 211(3):781-90, 1999. | ||
Latest revision as of 21:53, 10 December 2008
Home < BRAINSCutBack to NA-MIC Brains Collaboration
Summary
BRAINSCut is a software package for segmentation of structures using artificial neural networks. Currently this tool supports the segmentation of the following structures: brain, caudate, putamen, thalamus, hippocampus, anterior cerebellum, interior posterior cerebellum, superior posterior cerebellum, corpus medullary. Future regions will include the globus pallidus, amygdala, and nucleus accumbens. The command line uses the Slicer3 execution model framework.
Progress
- Integration with a high dimensional registration to the atlas probability map
- Improved thresholding of the output activation maps
- Code added to NITRC
- Coupled Neural network with MUSH Brain to generate a brain mask without requiring tissue classification
To Do
- Complete integration with the FANN library
- Link to a BSD style neural network library
- Look at the ability to use for segmentation of cortical regions
Key Investigators
- University of Iowa: Hans Johnson, Ronald Pierson, Kent Williams, Greg Harris, Vincent Magnotta
Figures
- BRAINSCut
Usage
BRAINSCut [--processinformationaddress <std::string>] [--xml] [--echo]
[--applyModel] [--trainModel] [--createVectors]
[--generateProbability] [--trainModelStartIndex <int>]
[--netConfiguration <std::string>] [--] [--version] [-h]
Description: Automatic Segmentation using neural networks
Author(s): Vince Magnotta, Hans Johnson, Greg Harris, Kent Williams
Where:
--processinformationaddress <std::string>
Address of a structure to store process information (progress, abort,
etc.). (default: 0)
--xml
Produce xml description of command line arguments (default: 0)
--echo
Echo the command line arguments (default: 0)
--applyModel
apply the neural net (default: 0)
--trainModel
train the neural net (default: 0)
--createVectors
create vectors for training neural net (default: 0)
--generateProbability
Generate probability map (default: 0)
--trainModelStartIndex <int>
Starting iteration for training (default: 0)
--netConfiguration <std::string>
XML File defining AutoSegmentation parameters
--, --ignore_rest
Ignores the rest of the labeled arguments following this flag.
--version
Displays version information and exits.
-h, --help
Displays usage information and exits.
Links
Papers
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. Neuroimage. 39(1):238-47, 2008.
- Magnotta VA, Heckel D, Andreasen NC, Cizadlo T, Corson PW, Ehrhardt JC, Yuh WT. Measurement of brain structures with artificial neural networks: two- and three-dimensional applications. Radiology. 211(3):781-90, 1999.