Difference between revisions of "2009 Winter Project Week LungImagingPlatform"
| (9 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2009_Winter_Project_Week|Project Week Main Page]] ]] | |[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2009_Winter_Project_Week|Project Week Main Page]] ]] | ||
| − | |[[]] | + | |[[Image:sagittalLungLobes.jpg]] |
| − | |[[]] | + | |[[Image:AirwayInspectorGUI_small.png]] |
|} | |} | ||
| Line 19: | Line 19: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
| − | Our goal is to develop a lung imaging platform for the clinical understanding of multiple lung diseases | + | Our goal is to develop a lung imaging platform for the clinical understanding of multiple lung diseases such as Chronic Obstructive Pulmonary Disease (COPD), Asthma, and Interstitial Lung Disease (ILD) among others. Quantitative lung imaging is a key component of on-going genetic and molecular biology studies that are being carried out at BWH and other centers across USA. A significant example of these new efforts is [[http://www.copdgene.org/|COPD Genetics Epidemiology]] multicenter study. The NAMIC Kit and Slicer 3 are ideal candidates for the implementation of such efforts. |
Our goals for this week are threefold: | Our goals for this week are threefold: | ||
| − | * | + | * Design the main platform components. |
* Dynamic programming approaches for the extraction of 3D airways. | * Dynamic programming approaches for the extraction of 3D airways. | ||
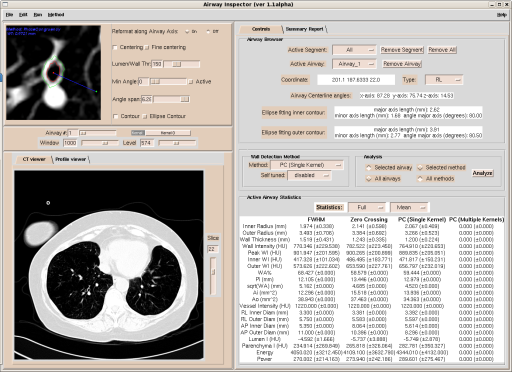
* Porting our current imaging platform, [[http://www.airwayinspector.org| Airway Inspector]] based on 3D Slicer to Slicer 3. | * Porting our current imaging platform, [[http://www.airwayinspector.org| Airway Inspector]] based on 3D Slicer to Slicer 3. | ||
| Line 31: | Line 31: | ||
<h1>Approach, Plan</h1> | <h1>Approach, Plan</h1> | ||
| + | Our approach is to develop a module in Slicer 3 that has the following components: | ||
| + | * A unique library that can be linked against and shared by other Slicer modules or external applications. | ||
| + | * Command Line Modules that encapsulate the individual algorithmic components of the platform without strings attached to a particular solution. The idea is to enable rapid prototyping and deployment of the solutions before they are fully integrated. | ||
| + | * Custom solutions for disease-oriented applications: our plan is to discuss how an application can customize the Slicer 3 layout. | ||
| + | We will focus on the following design aspects: | ||
| + | * MRML data structures for tubular-type anatomical structures. | ||
| + | * Support for quantitative imaging: handling data tables in Slicer 3 from MRML to Command Line Modules. | ||
| + | * ITK filter hierarchies for the main image analysis components: extraction of lung lobes, airways and vessels. | ||
| + | * Customizable GUI layout. | ||
</div> | </div> | ||
| Line 38: | Line 47: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| + | * LungImagingPlatform Module has been created. The code is currently available through the LMI repository (password = bwhspl): | ||
| + | :pserver:anonymous@cvs.spl.harvard.edu:/projects/cvs/slicer_lmi | ||
| + | * Slicer2 functionality has been ported to the module. | ||
| + | * The plan is to offer the module as an extension using the Extension infrastructure. This is still work in progress | ||
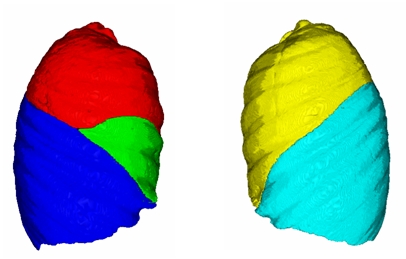
* A semiautomatic lobe segmentation has been successfully implemented in Slicer 3 as a Command Line Module. | * A semiautomatic lobe segmentation has been successfully implemented in Slicer 3 as a Command Line Module. | ||
| − | * Initial | + | * Design needs have been discussed and integration with vmtk for tubular network representation is going to be adopted. |
| − | + | * XNAT Desktop has been positively installed and tested as a platform to organize COPDGene datasets at BWH. Dicom rules have been defined to cover the needs of the project. | |
| + | * Initial discussions with Randy Golub have been carried out to leverage the Harvard XNAT enterprise resources. | ||
| + | * GUI needs have been discussed to enable a module-dependent GUI space with more flexible layouts for reporting visual and quantitative results. | ||
</div> | </div> | ||
| Line 48: | Line 63: | ||
</div> | </div> | ||
| − | ===References=== | + | === Related projects === |
| + | Other projects that share some common goals are: | ||
| + | * [http://www.na-mic.org/Wiki/index.php/2009_Winter_Project_Week_SlicerLayouts | User Interface Flexible Layouts] | ||
| + | * [http://www.na-mic.org/Wiki/index.php/2009_Winter_Project_Week_Slicer_VMTK | Vessel Segmentation in Slicer using VMTK ] | ||
| + | * [http://wiki.na-mic.org/Wiki/index.php/2009_Winter_Project_Week:GT_TubularSurfaceSeg | Tubular volumetric segmentation framework ] | ||
| + | |||
| + | === References === | ||
Latest revision as of 07:50, 9 January 2009
Home < 2009 Winter Project Week LungImagingPlatform Return to Project Week Main Page |
|
|
Key Investigators
- James Ross
- Raul San Jose
Objective
Our goal is to develop a lung imaging platform for the clinical understanding of multiple lung diseases such as Chronic Obstructive Pulmonary Disease (COPD), Asthma, and Interstitial Lung Disease (ILD) among others. Quantitative lung imaging is a key component of on-going genetic and molecular biology studies that are being carried out at BWH and other centers across USA. A significant example of these new efforts is [Genetics Epidemiology] multicenter study. The NAMIC Kit and Slicer 3 are ideal candidates for the implementation of such efforts.
Our goals for this week are threefold:
- Design the main platform components.
- Dynamic programming approaches for the extraction of 3D airways.
- Porting our current imaging platform, [Airway Inspector] based on 3D Slicer to Slicer 3.
Approach, Plan
Our approach is to develop a module in Slicer 3 that has the following components:
- A unique library that can be linked against and shared by other Slicer modules or external applications.
- Command Line Modules that encapsulate the individual algorithmic components of the platform without strings attached to a particular solution. The idea is to enable rapid prototyping and deployment of the solutions before they are fully integrated.
- Custom solutions for disease-oriented applications: our plan is to discuss how an application can customize the Slicer 3 layout.
We will focus on the following design aspects:
- MRML data structures for tubular-type anatomical structures.
- Support for quantitative imaging: handling data tables in Slicer 3 from MRML to Command Line Modules.
- ITK filter hierarchies for the main image analysis components: extraction of lung lobes, airways and vessels.
- Customizable GUI layout.
Progress
- LungImagingPlatform Module has been created. The code is currently available through the LMI repository (password = bwhspl):
:pserver:anonymous@cvs.spl.harvard.edu:/projects/cvs/slicer_lmi
- Slicer2 functionality has been ported to the module.
- The plan is to offer the module as an extension using the Extension infrastructure. This is still work in progress
- A semiautomatic lobe segmentation has been successfully implemented in Slicer 3 as a Command Line Module.
- Design needs have been discussed and integration with vmtk for tubular network representation is going to be adopted.
- XNAT Desktop has been positively installed and tested as a platform to organize COPDGene datasets at BWH. Dicom rules have been defined to cover the needs of the project.
- Initial discussions with Randy Golub have been carried out to leverage the Harvard XNAT enterprise resources.
- GUI needs have been discussed to enable a module-dependent GUI space with more flexible layouts for reporting visual and quantitative results.
Related projects
Other projects that share some common goals are:
- | User Interface Flexible Layouts
- | Vessel Segmentation in Slicer using VMTK
- | Tubular volumetric segmentation framework