Difference between revisions of "AHM2009:PNL"
(→Team) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 3: | Line 3: | ||
__NOTOC__ | __NOTOC__ | ||
==PNL Roadmap Project== | ==PNL Roadmap Project== | ||
| − | |||
{| | {| | ||
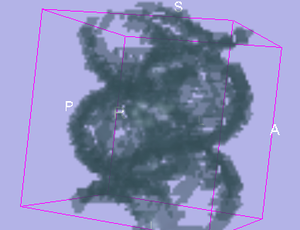
| − | |[[Image:Helix.png|thumb | + | |[[Image:Helix.png|thumb|Stochastic Tractography on helix phantom]] |
| − | |[[Image:Wmm.png|thumb | + | |[[Image:Wmm.png|thumb|White Matter Mask generated from phantom]] |
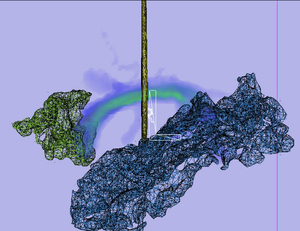
| − | |[[Image:STArcuate. | + | |[[Image:STArcuate.png|thumb|Stochastic Tractography of Arcuate Fasciculus]] |
| − | |[[Image:Step1.png|thumb| | + | |[[Image:Step1.png|thumb|56px|Stochastic Tractography Module]] |
| − | |[[Image:Step2.png|thumb| | + | |[[Image:Step2.png|thumb|152px|Stochastic Tractography Module]] |
| − | |||
|} | |} | ||
== Overview == | == Overview == | ||
| − | ;* What problem does the pipeline solve? | + | ;*What problem does the pipeline solve? |
Most tractography methods (Deterministic/Principal Diffusion Direction) estimate fibers by tracing the maximum direction of diffusion. | Most tractography methods (Deterministic/Principal Diffusion Direction) estimate fibers by tracing the maximum direction of diffusion. | ||
| Line 32: | Line 30: | ||
Functionality of Python Stochastic Tractography module in Slicer 3.0 includes: | Functionality of Python Stochastic Tractography module in Slicer 3.0 includes: | ||
| − | *Preprocessing | + | ;*Preprocessing |
Reading DWI and ROI files in nhdr format | Reading DWI and ROI files in nhdr format | ||
| Line 44: | Line 42: | ||
Producing diffusion indices (FA, Mode, Trace) | Producing diffusion indices (FA, Mode, Trace) | ||
| − | *Creating probability maps (parameters involve number of tracts per seed, tract length, step size, stopping criteria) | + | ;*Creating probability maps (parameters involve number of tracts per seed, tract length, step size, stopping criteria) |
Probability maps can be saved as ROIs, and either used directly, or thresholded (at certain probability, step claimed by few publications to remove noise) in slicer to mask and compute average FA, Mode, Trace for entire connection. Diffusion indices can be also weighted by the probability of connection for each voxel. | Probability maps can be saved as ROIs, and either used directly, or thresholded (at certain probability, step claimed by few publications to remove noise) in slicer to mask and compute average FA, Mode, Trace for entire connection. Diffusion indices can be also weighted by the probability of connection for each voxel. | ||
| Line 66: | Line 64: | ||
==Team== | ==Team== | ||
| − | |||
| − | |||
| − | |||
| − | + | {| | |
| − | + | |[[Image:Logo_pnl2.png|75px]] | |
| − | + | |valign="top"| DBP: Marek Kubicki, MD, PhD, Sylvain Bouix, PhD, Julien De Siebenthal, PhD, Doug Terry, BS, | |
| − | [[Image:csail.jpg| | + | [http://pnl.bwh.harvard.edu/ Psychiatry Neuroimaging Laboratory] |
| − | Core 1: Polina Golland, MIT | + | |- |
| − | + | |[[Image:csail.jpg|75px]] | |
| − | + | |valign="top"|Core 1: Polina Golland, [http://www.csail.mit.edu/ CSAIL MIT] | |
| − | + | |- | |
| − | + | |[[Image: Kitware.png|75px]] | |
| − | [[Image: Kitware.png| | + | |valign="top"| Core 2: Brad Davis, Steve Pieper, [http://www.kitware.com/ Kitware] |
| − | Core 2: Brad Davis, Steve Pieper, Kitware | + | |} |
==Outreach== | ==Outreach== | ||
Latest revision as of 17:33, 17 February 2009
Home < AHM2009:PNL
PNL Roadmap Project
Overview
- What problem does the pipeline solve?
Most tractography methods (Deterministic/Principal Diffusion Direction) estimate fibers by tracing the maximum direction of diffusion.
A limitation of this approach is that, in practice, several factors introduce uncertainty in the tracking procedure, including, noise, splitting and crossing fibers, head motion and image artifacts.
Stochastic tractography attempts to quantify the uncertainty associated with estimated fibers, by using a propagation model based on stochastics and regularization, which allows paths originating at one point to branch and return a probability distribution of possible paths.
Based on probability functions, using a sequential importance sampling technique (Bjornemo et al., 2002), one can generate thousands of fibers starting in the same point by sequentially drawing random step directions. This gives a very rich model of the fiber distribution, as contrasted with single fibers produced by conventional tractography methods. Moreover, from a large number of sampled paths, probability maps can be generated, providing better estimates of connectivity between several anatomical locations.
- Who is the targeted user?
Since diffusion direction uncertainty within the gray matter is quite significant; principal diffusion direction approaches usually do not work for tracking between two gray matter regions. Thus if one requires finding connections between a priori selected anatomical gray matter regions, defined either by anatomical segmentations (in case of using structural ROI data), or functional activations (in case of megring DTI with fMRI), stochastic tractography seems to be the method of choice. Stochastic Tractography is also comparable, if not better, in defining large white matter fiber bundles, especially those traveling through white matter regions characterized by increased diffusion uncertainty (fiber crossings).
- How does the pipeline compare to state of the art?
To date, there are two popular toolboxes that include probabilistic/stochastic tractography modules. Functionality of both modules (FSL and PICO) is limited, and software is not open source.
Detailed Information about the Pipeline
Stochastic Tractography pipeline has been written in Python, and is part of current Slicer3 release.
Functionality of Python Stochastic Tractography module in Slicer 3.0 includes:
- Preprocessing
Reading DWI and ROI files in nhdr format
Smoothing DWI data
Creating brain and white mater masks
Removing artifacts in WM masks by comparing this mask with FA map
Producing diffusion indices (FA, Mode, Trace)
- Creating probability maps (parameters involve number of tracts per seed, tract length, step size, stopping criteria)
Probability maps can be saved as ROIs, and either used directly, or thresholded (at certain probability, step claimed by few publications to remove noise) in slicer to mask and compute average FA, Mode, Trace for entire connection. Diffusion indices can be also weighted by the probability of connection for each voxel.
- Parameters/steps that need to be adjusted using someone else's data
Software was tested on 3 sets of data (1.5T anisotropic Siemens, 3T anisotropic GE, 3T isotropic GE), and works for all of them without major modifications.
Software & documentation
- Download Slicer 3.0 here.
- Module documentation can be found here:
- Software that you will also need to launch:
Team

|
DBP: Marek Kubicki, MD, PhD, Sylvain Bouix, PhD, Julien De Siebenthal, PhD, Doug Terry, BS, |

|
Core 1: Polina Golland, CSAIL MIT |
| Core 2: Brad Davis, Steve Pieper, Kitware |
Outreach
- Visit our publication database.
- Planned outreach activities (including presentations, tutorials/workshops) at conferences
- 1. Method of stochastic tractography, along with the module functionality will be presented at the DTI symposium during World Biological Psychiatry Symposium in Florence, Italy in April 2009.
- 2. Clinical applications, including results of arcuate fasciculus study in schizophrenia will be presented at the Biological Psychiatry Congress in Vancouver, Canada in May 2009.