Difference between revisions of "CTSC Ellen Grant, CHB"
| (128 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
=Mission= | =Mission= | ||
| + | |||
| + | =Use-Case Goals= | ||
| + | We will approach this use-case in three distinct steps, including Basic Data Management, Query Formulation and Processing Support. | ||
| + | * '''Step 1: Data Management''' | ||
| + | ** '''Step 1a.:''' Describe and upload retrospective datasets (roughly 1 terabyte) onto the CHB XNAT instance and confirm appropriate organization and naming scheme via web GUI. | ||
| + | ** '''Step 1b.:''' Describe and upload new acquisitions as part of data management process. | ||
| + | * '''Step 2: Query Formulation''' | ||
| + | ** making specific queries using XNAT web services, | ||
| + | ** data download conforming to specific naming convention and directory structure, using XNAT web services | ||
| + | ** ensure all queries required to support processing workflow are working. | ||
| + | * '''Step 3: Data Processing''' | ||
| + | ** Implement & execute the script-driven tractography workflow using web services, | ||
| + | ** describe and upload results. | ||
| + | ** ensure results are appropriately structured and named in repository, and queriable via web GUI and web services. | ||
| Line 12: | Line 26: | ||
* clinicians | * clinicians | ||
* IT staff | * IT staff | ||
| + | |||
| + | =Outcome Metrics= | ||
| + | '''Step 1: Data Management''' | ||
| + | * Visual confirmation (via web GUI) that all data is present, organized and named appropriately | ||
| + | * other? | ||
| + | '''Step 2: Query Formulation''' | ||
| + | * Successful tests that responses to XNAT queries for all MRIDs given a protocol name match results returned from currently-used search on the local filesystem. | ||
| + | * Query/Response should be efficient | ||
| + | '''Step 3: Data Processing''' | ||
| + | * Pipeline executes correctly | ||
| + | * Pipeline execution not substantially longer than when all data is housed locally | ||
| + | * other? | ||
| + | '''Overall''' | ||
| + | * Local disk space saved? | ||
| + | * Data management more efficient? | ||
| + | * Data management errors reduced? | ||
| + | * Barriers to sharing data lowered? | ||
| + | * Processing time reduced? | ||
| + | * User experience improved? | ||
| + | |||
| + | = Fundamental Requirements = | ||
| + | * System must be accessible 24/7 | ||
| + | * System must be redundant (no data loss) | ||
| + | * Need a better client than current web GUI provides: | ||
| + | ** faster | ||
| + | ** PACS-like interface. | ||
| + | ** image viewer should open in SAME window (not pop up a new) | ||
| + | ** number of clicks to get to image view should be as few as possible. | ||
| + | |||
| + | =Outstanding Questions = | ||
| + | |||
| + | Plans for improving web GUI? | ||
=Data= | =Data= | ||
| − | There are currently about | + | Retrospective data consists of ~1787 studies, ~1TB total. |
| − | * | + | Data consists of |
| − | * | + | * MR data, DICOM format |
| − | + | * Demographics from DICOM headers | |
| − | = | + | * Subsequent processsing generates ".trk" files |
| + | * ascii text files ".txt" | ||
| + | * files that contain protocol information | ||
| + | |||
| + | = Progress notes = | ||
| + | ==Sprint 2== | ||
| + | * '''Meeting on 4/08/2010:''' work has restarted on this effort. In the April/May timeframe: | ||
| + | ** Wendy will revisit the tcl scripts and ensure that they are compliant with NEW XNAT 1.4 release. | ||
| + | ** Rudolph and Dan Ginsburg will choose and provide three datasets for upload with these scripts. | ||
| + | ** Wendy will upload these datasets using the previously developed mapping between Rudolph's data description and XNAT's schema. | ||
| + | ** Wendy will send Dan a pointer to XNAT 1.4 documentation and tutorials.(Done: see [http://www.na-mic.org/Wiki/index.php/CTSC_Ellen_Grant,_CHB#Helpful_XNAT_links here] ) | ||
| + | ** After this upload, we will all visit and review organization of data on XNAT. | ||
| + | ** Dan will also abstract their XML description of metadata used to populate their web-based front end, to make a friendlier adaptor for XNAT schema. | ||
| + | ** Once approach for organizing and managing data on XNAT is settled, Wendy will begin writing scripts to | ||
| + | *** make appropriate queries, and | ||
| + | *** translate xml responses into appropriate format for web-based front end to display. | ||
| + | |||
| + | === XNAT Evaluation - Dan G === | ||
| + | * Installed XNAT 1.4 in Ubuntu 9.10 VM. Got tripped up a bit on issue that was solved on xnat_discussion list [http://groups.google.com/group/xnat_discussion/browse_thread/thread/a400f8347118cbe4 link] | ||
| + | * Can't get user sessions to work, so I modified Wendy's scripts to pass -u/-p on each call, awaiting fix on xnat_discussion list [http://groups.google.com/group/xnat_discussion/browse_thread/thread/fefd777638a8ad0d link] | ||
| + | * Next step is to go through xnat.xsd schema and see how it maps to the fields we are currently using in our own XML. Will post results on Wiki when done. | ||
| + | |||
| + | === XNAT Schema Mapping - Dan G === | ||
| + | I went through the exercise of looking at the data that we currently use in our custom XML to see if/where it could be found in the XNAT XML Schema. First, here is a sample entry from our XML: | ||
| + | |||
| + | |||
| + | <PatientRecord> | ||
| + | <PatientID>...</PatientID> | ||
| + | <Directory>...</Directory> | ||
| + | <PatientName>....</PatientName> | ||
| + | <PatientAge>...</PatientAge> | ||
| + | <PatientSex>...</PatientSex> | ||
| + | <PatientBirthday>...</PatientBirthday> | ||
| + | <ImageScanDate>...</ImageScanDate> | ||
| + | <ScannerManufacturer>...</ScannerManufacturer> | ||
| + | <ScannerModel>...</ScannerModel> | ||
| + | <SoftwareVer>...</SoftwareVer> | ||
| + | <Scan>... </Scan> | ||
| + | <Scan>... </Scan> | ||
| + | <Scan>... </Scan> | ||
| + | <Scan>.... </Scan> | ||
| + | <User>...</User> | ||
| + | <Group>...</Group> | ||
| + | </PatientRecord> | ||
| + | |||
| + | The following table maps between each of these tags and the xnat.xsd schema in XNAT 1.4: | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Tag | ||
| + | ! XNAT Schema Equivalent | ||
| + | |- | ||
| + | | PatientID | ||
| + | | xnat:subjectData/label (this is where Wendy's script stores the MRID), retrieved with (e.g., ./XNATRestClient -u USERNAME -p PASSWORD -host http://localhost:8080/xnat/ -m GET -remote /REST/projects/FNNDSC_test/subjects) | ||
| + | |- | ||
| + | | Directory | ||
| + | | xnat:experiment/label | ||
| + | |- | ||
| + | | PatientName | ||
| + | | xnat:imageSessionData/dcmPatientName | ||
| + | |- | ||
| + | | PatientAge | ||
| + | | xnat:demographicData/xnat:abstractDemographicData/(dob,yob,age) | ||
| + | |- | ||
| + | | PatientSex | ||
| + | | xnat:demographicData/gender | ||
| + | |- | ||
| + | | PatientBirthday | ||
| + | | xnat:demographicData/xnat:abstractDemographicData/(dob,yob,age) | ||
| + | '''Q''': I think dob/yob/age are mutually exclusive, so if age is put in I am not sure if DOB is available? | ||
| + | |- | ||
| + | | ImageScanDate | ||
| + | | xnat:experimentData/date | ||
| + | |- | ||
| + | | ScannerManufacturer | ||
| + | | xnat:imageSessionData/scanner/manufacturer | ||
| + | NOTE: It looks like there might be server config to have the drop-down have pre-defined values | ||
| + | |- | ||
| + | | ScannerModel | ||
| + | | xnat:imageSessionData/scanner/model | ||
| + | |- | ||
| + | | SoftwareVer | ||
| + | | There are a lot of useful fields in xnat:mrScanData, but I did not in particular find the software version. | ||
| + | '''Q''': Should it be added? Did I miss it? | ||
| + | |- | ||
| + | | Scan | ||
| + | | xnat:mrScanData/parameters/scanSequence | ||
| + | |- | ||
| + | | User, Group | ||
| + | | I don't think these belong in here once we go to XNAT, we will have to re-evaluate how we do authentication. Ideally we should use the accounts in XNAT rather than what we do now. | ||
| + | |} | ||
| + | |||
| + | ==Sprint 1== | ||
| + | * Trying experiment with Rudolph -- review of test data, and trial run of bulk upload script | ||
| + | * Wendy is working on pydicom script to optionally anonymize from this script | ||
| + | * and also working on pydicom script to parse headers and add "fileData" on upload | ||
| + | * also working on extending the XNATquery.tcl tool to perform flexible query/parse. | ||
| + | * Rudolph can have a look at the testdata upload on central & discuss with wendy | ||
| + | * if ok, rudolph can try upload script on central, or on chb instance (may be unexpected differences?) | ||
| + | |||
| + | =Use Case Assessment #2= | ||
| + | |||
| + | == Helpful XNAT links == | ||
| + | |||
| + | [http://nrg.wikispaces.com/XNAT XNAT 1.4 documentation] | ||
| + | |||
| + | [http://nrg.wikispaces.com/Understanding+the+XNAT+Data+Model Data Model] | ||
| + | |||
| + | [http://central.xnat.org/schemas/xnat/xnat.xsd XSD document ] | ||
| + | |||
| + | [http://nrg.wikispaces.com/XNAT+REST+API REST API (the API we'll use)] | ||
| + | |||
| + | [http://nrg.wikispaces.com/XNAT+REST+API+Usage REST API usage guide ] | ||
| + | |||
| + | [http://nrg.wikispaces.com/XNAT+REST+Quick+Tutorial REST tutorial ] | ||
| + | |||
| + | [http://nrg.wikispaces.com/XNAT+REST+XML+Path+Shortcuts XML path shortcuts & xsiTypes] | ||
| + | |||
| + | =Use Case Assessment #1= | ||
| + | |||
| + | == Current Data Management Process== | ||
| + | |||
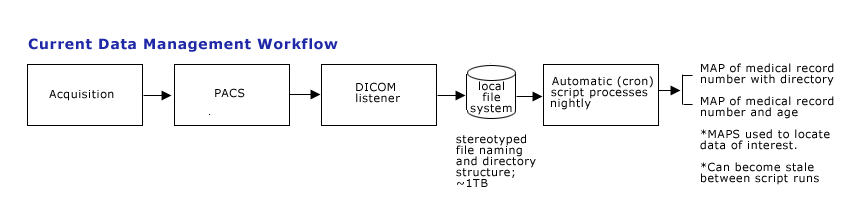
| + | DICOM raw images are produced at radiology PACS at MGH, and are manually pushed to the PACS hosted on KAOS resided at MGH NMR center. The images are processed by a set of PERL scripts to be renamed and re-organized into a file structure where all images for a study are saved into a directory named for the study. DICOM images are currently viewed with Osiris on Macs. | ||
| + | |||
| + | [[image:CTSCInformatics_GrantPienaarCurrentDataManagement.png]] | ||
| + | |||
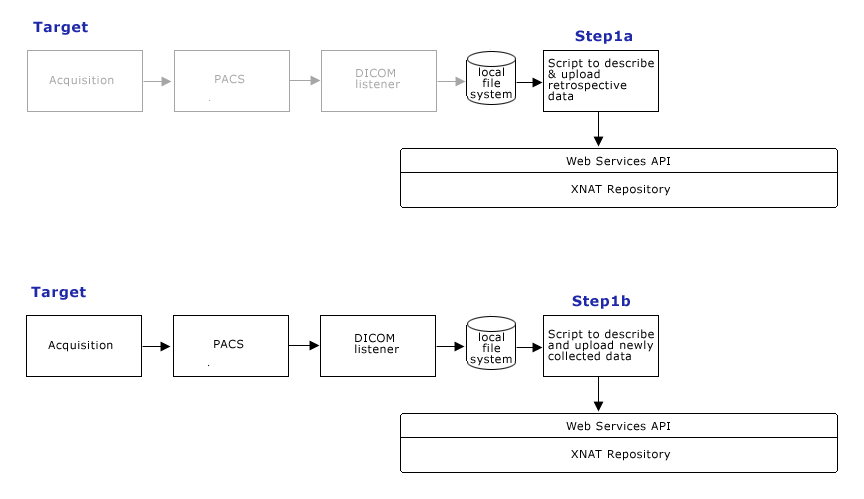
| + | == Target Data Management Process (Step 1) == | ||
| + | '''Step 1''': | ||
| + | Develop an Image Management System for BDC (IMS4BDC) with which at least the following can be done: | ||
| + | * Move images from MGH (KAOS) to a BDC machine at Children's | ||
| + | * '''Step 1a:''' Import legacy data into IMS4BDC from existing file structure and CDs | ||
| + | * '''Step 1b:''' Write scripts to execute upload of newly acquired data. | ||
| + | |||
| + | [[image:CTSCInformatics_GrantPienaarDataManagementStep1.png]] | ||
| + | |||
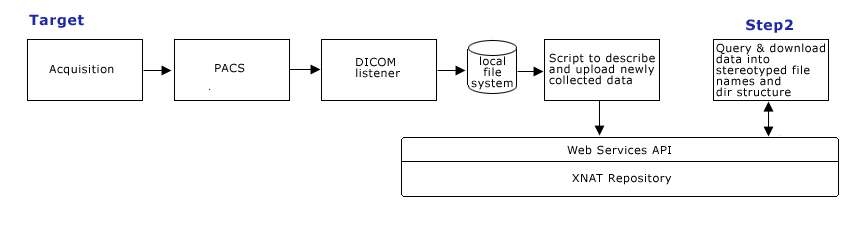
| + | == Target Query Formulation (Step 2) == | ||
| + | '''Step 2.''' Develop Query capabilities using scripted client calls to XNAT web services, such as: | ||
| + | |||
| + | Show all subjectIDs scanned with protocol_name = ProtocolName | ||
| + | Show all diffusion studies where patients ages are < 6 | ||
| + | |||
| + | * Scripting capabilities: Scripts need to query and download data into appropriate directory structure, and support appropriate naming scheme to be compatible with existing processing workflow. | ||
| + | |||
| + | [[image:CTSCInformatics_GrantPienaarDataManagementStep2.png]] | ||
| + | |||
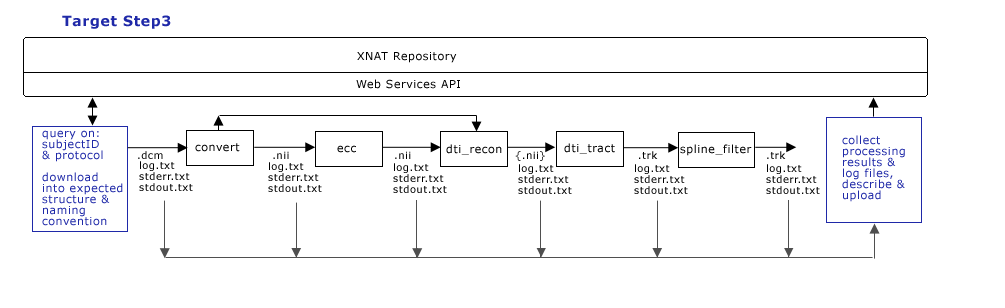
| + | == Target Processing Workflow (Step 3) == | ||
| + | '''Step 3:''' | ||
| + | |||
| + | [[image:CTSCInformatics_GrantPienaarDataManagementStep3.png]] | ||
| + | |||
| + | * Execute query/download scripts | ||
| + | * Run processing locally, on cluster, etc. | ||
| + | * Describe & upload processing results | ||
| + | * (eventually want to) Share images with clinical physicians | ||
| + | * (eventually want to) Export post-processed data back to clinical PACS | ||
| + | |||
| + | = Fitting Data to XNAT Data Model = | ||
| + | |||
| + | ==Test data from Rudolph== | ||
| + | |||
| + | I think we have this mappint from project to XNE data model: | ||
| + | * MRID = SubjectID (1687 subjects?) | ||
| + | * each SubjectID may have single experiment, but multiple MRSessions within that experiment | ||
| + | * each "storage" directory for a particular MRID (in dcm_mrid.log) = MRSessionID | ||
| + | * each scan listed (int toc.txt) = ScanID in the MRsession | ||
| + | * important metadata contained in dicom headers, in dcm_mrid_age.log, dcm_mrid_age_days.log, and in the toc.txt file in each session directory. | ||
| + | |||
| + | This gives us a unique way to | ||
| + | * have unique subject IDs | ||
| + | * have unique MRSessionIDs for each subject, | ||
| + | * have unique scanIDs within each MRSession | ||
| + | * search for subject (by ID, age, or dicom header info) or | ||
| + | * search for image data by age (or dicom header info) | ||
| + | |||
| + | === As regards anonymization === | ||
| + | Rudolph doesn't specifically need XNAT to do the anonymization. Wants XNAT to contain all relevant data and where/if necessary export/transmit DICOM data anonymized. | ||
| + | |||
| + | Rudolph has own MatLAB script that can do batch anonymization -- but if possible XNAT package should probably provide a means for that. | ||
| + | |||
| + | === Draft approach to uploading data for Rudolph === | ||
| + | * Create a Project on the webGUI | ||
| + | * Write a webservices-client script that will batch: | ||
| + | ** create subject (tested) | ||
| + | ** create experiment for subject (tested) | ||
| + | ** create mrsession for subject (tested) | ||
| + | ** for each scan in mrsession | ||
| + | *** anonymize (not sure) | ||
| + | *** do dicom markup (not sure) | ||
| + | *** add other metadata from toc.txt and *.log files (not sure) | ||
| + | *** upload scan data into db (tested) | ||
| + | |||
| + | |||
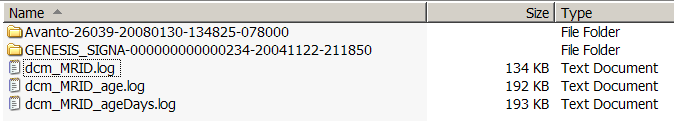
| + | === Current data (subset) organization === | ||
| + | [[image:RudolphTestData.png]] | ||
| + | |||
| + | The top level directory contains | ||
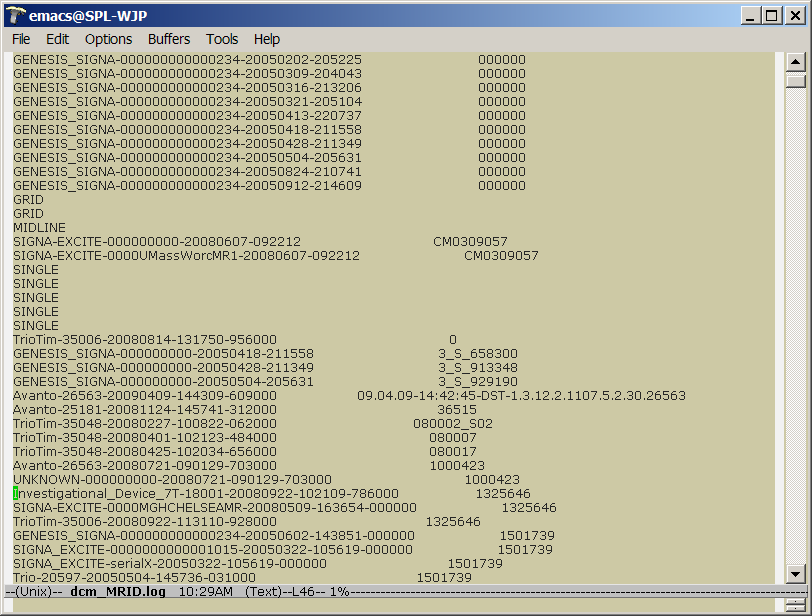
| + | * a dcm_MRID.log file that contains a mapping between MRID's (PatientIDs?) and unique MRSessionNames | ||
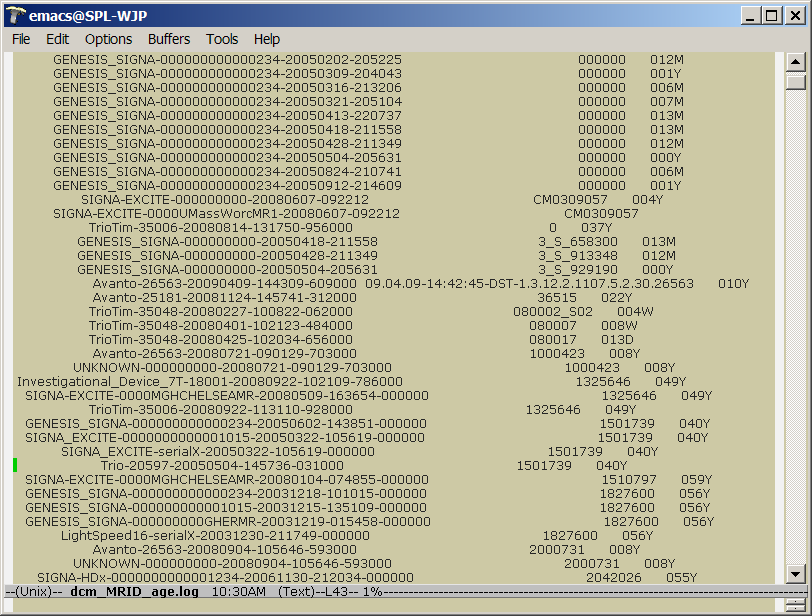
| + | * a dcm_MRID_age.log file that maps MRID's to ages in months and years | ||
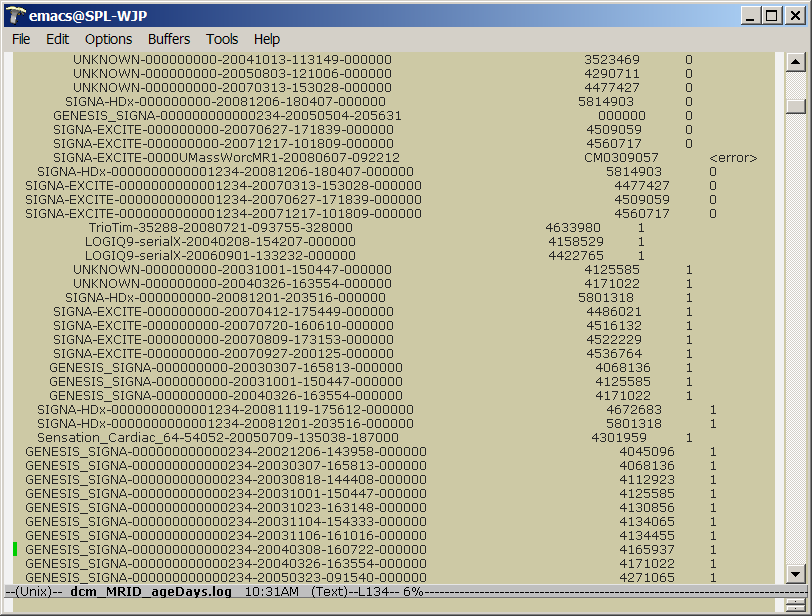
| + | * a dcm_MRID_age_days.log file tha tmaps MRID's to ages in days | ||
| + | * subdirectories named for MRSessions. | ||
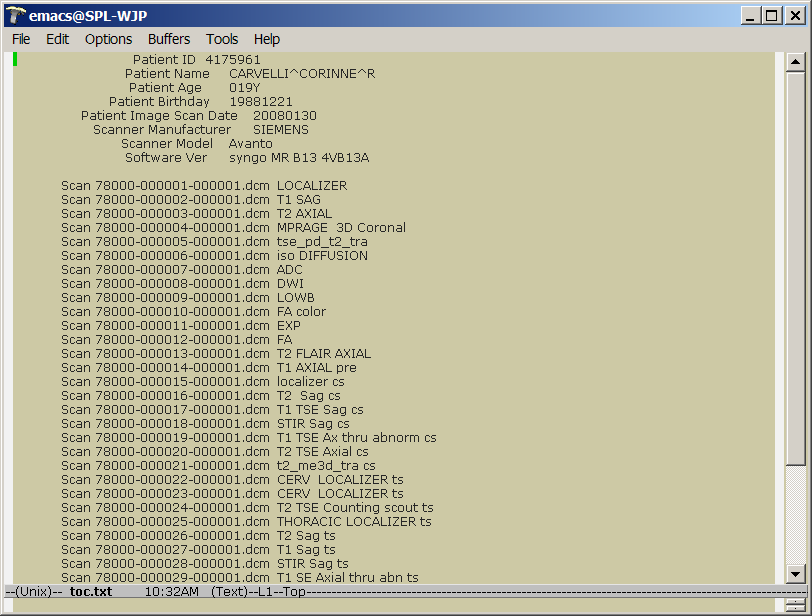
| + | * each subdirectory contains a toc.txt file that includes patient and session information and a list of scans and scan types. | ||
| + | |||
| + | See examples below: | ||
| + | |||
| + | [[image:dcm_MRID_log.png | dcm_MRID.log]] | ||
| + | |||
| + | [[image:dcm_MRID_age_log.png | dcm_MRID_age.log]] | ||
| + | |||
| + | [[image:dcm_MRID_age_days_log.png | dcm_MRID_age_days.log]] | ||
| + | |||
| + | [[image:RudolphSession_toc_txt.png | toc.txt]] | ||
| + | |||
| + | ==Questions sent to Rudolph about test data:== | ||
| + | |||
| + | First, in the top-level dir, there are three log files: | ||
| + | dcm_MRID.log | ||
| + | dcm_MRID_age.log | ||
| + | dcm_MRID_ageDays.log | ||
| + | |||
| + | 1. do these files contain the MRIDs for *all* subjects in the entire | ||
| + | retrospective study? | ||
| + | |||
| + | --Yes, at least current to the timestamp of the log file. | ||
| + | |||
| + | 2. Some MRIDs appear to be purely numerical, and some | ||
| + | alphanumerical. (3_S_658300). Is that correct? | ||
| + | |||
| + | Yes again. Unfortunately there seems to no standard technique for spec'ing | ||
| + | the MRID number. This number, however, is the key most often used by clinicians, | ||
| + | and thus is a primary key for the database. Problems abound, of course -- the MRID is | ||
| + | linked to a single patient, but is not necessarily guaranteed to be unique. A patient | ||
| + | keeps the same MRID, so multiple scans result in multiple instances. The combination of | ||
| + | MRID+<storageDirectory> is unique (but also redundant, since the <storageDirectory> is | ||
| + | unique, by definition. So essentially the log files are lookup tables for MRIDs and | ||
| + | actual storage locations in the filesystem. | ||
| + | |||
| + | 3. Two age files, one contains age in months or years (1687 entries) -- the other | ||
| + | contains age in days (1687 entries): | ||
| + | * does this mean there are 1687 MRID's in total? | ||
| + | * what does age (days) = -1 mean? | ||
| + | |||
| + | These are just convenience files. Often times a typical 'query' would be: | ||
| + | "findallMRID WHERE age IN <someAgeConstraint> AND protocol IN <someProtocolConstraint>." | ||
| + | |||
| + | The dcm_MRID_age.log maps the MRID to a storage location, and provides the | ||
| + | age as tagged in the DICOM header. Of course, mixing different age formats | ||
| + | (like 012M and 004W etc) isn't batch processing friendly. So, the | ||
| + | dcm_MRID_ageDays.log converts all these age specifiers to days, and sorts | ||
| + | the table on that field. | ||
| + | |||
| + | The '-1' means that some error occurred in the day calculation. | ||
| + | Most likely, the associated age value wasn't present. | ||
| + | |||
| + | 4. And the two dataset directories you shared: | ||
| + | |||
| + | Avanto-26039-20080130-134825-078000/ | ||
| + | |||
| + | GENESIS_SIGNA-000000000000234-20041122-211850/ | ||
| + | |||
| + | each directory contain data and a .toc file that includes: | ||
| + | |||
| + | * the "PatientID" is this equivalent to MRID? | ||
| + | * and some other info including age, scan date, etc. | ||
| + | * the filenames and scan types of a *set* of scans: | ||
| + | ** collected in one MRsession on that scan date? | ||
| + | ** or in the entire retrospective project? | ||
| + | * and is all the data for the set of scans listed contained in this directory? | ||
| + | |||
| + | true: PatientID == MRID | ||
| + | The filenames and scantypes correspond to one session on that scan date. | ||
| + | Other scan dates for that MRID will be in different directories. | ||
| + | Essentially, the data is packed according to | ||
| + | <scannerSpec>-<scanprefix>-<scandate>-<scantime>-<trailingID>. | ||
| + | |||
| + | ==Upload tests:== | ||
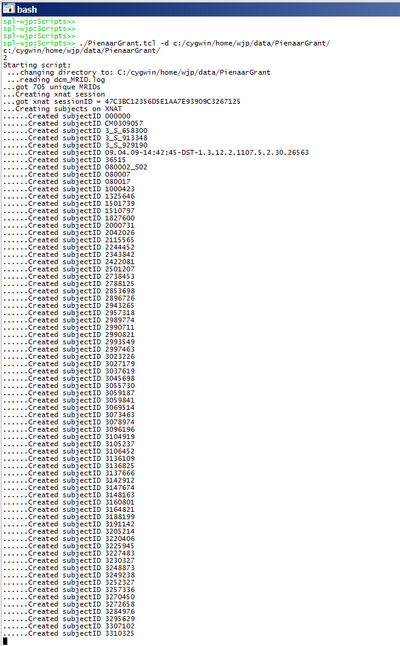
| + | Remote script to upload -- parsing Rudolph's data and making webservices calls to create subjects: | ||
| + | |||
| + | [[image:PG_WS_createsubjects.png|thumb|400px|center|subject creation while running; each subject takes ~ 1second]] | ||
| + | |||
| + | = Local data organization to XNAT data model mapping = | ||
| + | implementing script to batch upload data from local to remote using webservices: | ||
| + | |||
| + | <pre> | ||
| + | #---------------------------------------------------------------------------------------- | ||
| + | # Local and Remote data organization notes | ||
| + | #---------------------------------------------------------------------------------------- | ||
| + | # Batch upload of retrospective study maps local data organization | ||
| + | # to XNAT data model in the following way: | ||
| + | # xnat Project = PienaarGrant | ||
| + | # xnat Project contains list of unique xnat SubjectIDs (= local MRIDs) | ||
| + | # Each xnat SubjectID contains a list of xnat "Experiments" | ||
| + | # Each xnat Experiment is an "MRSession" with unique ID (and label = local dirname from dcm_mrid.txt) | ||
| + | # Each xnat MRSession labeled with local dirname contains set of xnat "Scans" (listed in local toc.txt) | ||
| + | # Each xnat Scan contains the image data. | ||
| + | # Project, Subjects, Experiments, and Scans can all have searchable metadata | ||
| + | # Projects, Experiments can have associated files (dcm_mrid.txt, toc.txt, error logs, etc.) | ||
| + | |||
| + | |||
| + | #---------------------------------------------------------------------------------------- | ||
| + | # A schematic of the local organization looks like this: | ||
| + | #---------------------------------------------------------------------------------------- | ||
| + | # | ||
| + | # Project root dir | ||
| + | # | | ||
| + | # { MRSession1, MRSession2, ..., MRSessionN} + {projectfiles = dcm_MRID.log, dcm_MRID_age.log, dcm_MRID_ageDays.log, error.log} | ||
| + | # |... | ||
| + | # { scanfiles=scan1.dcm, scan2.dcm,...,scanM.dcm } + {sessionfiles = toc.txt, toc.err, log/*, log_V/*} | ||
| + | # | ||
| + | |||
| + | |||
| + | #---------------------------------------------------------------------------------------- | ||
| + | # A schematic of the xnat organization looks like this: | ||
| + | #---------------------------------------------------------------------------------------- | ||
| + | # | ||
| + | # Project + {projectfiles.zip} | ||
| + | # | | ||
| + | # { SubjectID1, SubjectID2, ... SubjectIDN } | ||
| + | # ...| | ||
| + | # List of "Experiments" { MRSession, MRSession, ..., MRSession} | ||
| + | # ...| | ||
| + | # {scan1...scanN} + {sessionfiles.zip} | ||
| + | # | ||
| + | # An XNAT "Experiment" is an XNAT "imagingSession" | ||
| + | # An ImagingSession may be a MRSession, PETSession or CTSession (on central.xnat.org) | ||
| + | # -we are calling each local directory for a given MRID a new XNAT Experiment (MRSession). | ||
| + | # Each XNAT "imagingSession" contains a collection of XNAT "scans". | ||
| + | # we are calling each scan in a given local directory an XNAT "scan" in the MRSession. | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | = Experiment to try with Rudolph = | ||
| + | |||
| + | * Upload script is ready to be tested. | ||
| + | * Query and Download script still being developed | ||
| + | |||
| + | == Resources needed to run the scripts == | ||
| + | * [http://sourceforge.net/projects/tcl/files/ Tcl (any recent version will do)] | ||
| + | * [http://nrg.wustl.edu/1.4/xnat_tools.zip XNAT's Rest Client (with XNAt tools)] | ||
| + | * [http://code.google.com/p/pydicom/downloads/list pydicom (not needed yet!)] | ||
| + | |||
| + | == Scripts (ready to test for bulk upload) == | ||
| + | * [[media:PienaarGrantLabScripts.zip | Tcl scripts (zip file) ]] | ||
| + | |||
| + | == Instructions & Notes == | ||
| + | |||
| + | Rudolph: | ||
| − | + | Results of this script's upload are on central.xnat.org under the test project "PienaarGrantRetrospectiveTest". This is a private project because it contains protected medical information. | |
| − | + | * once you make an account on central, send me your login and I'll add you to the list of users who can access the data. | |
| + | * Then, click thru and let me know if the way data is organized appropriately matches your data. | ||
| + | * I've only uploaded the data for mrid=000000, and only one mrsession was in the test set -- so only one MRSession has scan data in it: | ||
| + | ** GENESIS_SIGNA-000000000000234-20041122-211850 | ||
| + | * if this doesn't work for you, let's meet and figure out how to fix. | ||
| + | * also, i've only tested for mrid=000000, and not for all mrids, so don't know whether there's a surprise when the code is run for all mrids in dcm_MRID.log. | ||
| + | * script is intended to work for initial bulk upload and for incremental uploads too (tho probably inefficiently) -- adding new experiments and their data as they appear in new dcm_MRID.log and other files. We'll have to test this. | ||
| − | = | + | === to use: === |
| − | + | * Create a test project using xnat web gui | |
| + | * Using the "Manage" tab: | ||
| + | ** Choose to make the project "private" | ||
| + | ** Choose to place data directly into the databasee (not prearchive) | ||
| + | ** Choose to skip the quarantine | ||
| + | * Download the XnatRESTClient (link above in 'resources' section) | ||
| + | * Download and install Tcl (link above in 'resources' section) | ||
| + | * Download the scripts and unzip (link above) | ||
| + | * Edit the XNATglobals.tcl file to customize | ||
| + | * run ./PGBulkUp.tcl -d rootDataDirectory | ||
| + | * the script will spew lots of text. | ||
| + | * some error messages pop up from time to time, and I'm not sure what causes the periodic croak: | ||
| + | ** ...>>>>>>ERROR: AddScanData got unexpected response from remote PUT of scan data: The scan may not have been properly created. Perform QA check for: subject=000000 MRSession=CENTRAL_E00702 scan=Scan4 on remote host. | ||
| + | ** ...>>>>>>ERROR: Got error response from XNAT host. | ||
| + | ** ...>>>>>>ERROR: Got error message instead of experimentID from remote PUT of experiment: Experiment may not have been properly created. Please perform QA check for 000000 on remote host. | ||
| + | * So we should save the script output to see whether problems pop up. | ||
| − | + | =PROJECT STATUS= | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * 9/29/09 - Scripts for bulk uploading using web services client made available for testing. | |
| + | ** Right now these scripts will upload entire retrospective study | ||
| + | ** They do not parse DICOM headers and add header information to XNAT's "fileData" | ||
| + | ** They do not anonymize | ||
| − | + | This seems like a reasonable first step since rudolph has his own scripts to anonymize, but did not want his data necessarily anonymized in the repository. | |
Latest revision as of 18:35, 15 April 2010
Home < CTSC Ellen Grant, CHBBack to CTSC Imaging Informatics Initiative
Contents
- 1 Mission
- 2 Use-Case Goals
- 3 Participants
- 4 Outcome Metrics
- 5 Fundamental Requirements
- 6 Outstanding Questions
- 7 Data
- 8 Progress notes
- 9 Use Case Assessment #2
- 10 Use Case Assessment #1
- 11 Fitting Data to XNAT Data Model
- 12 Local data organization to XNAT data model mapping
- 13 Experiment to try with Rudolph
- 14 PROJECT STATUS
Mission
Use-Case Goals
We will approach this use-case in three distinct steps, including Basic Data Management, Query Formulation and Processing Support.
- Step 1: Data Management
- Step 1a.: Describe and upload retrospective datasets (roughly 1 terabyte) onto the CHB XNAT instance and confirm appropriate organization and naming scheme via web GUI.
- Step 1b.: Describe and upload new acquisitions as part of data management process.
- Step 2: Query Formulation
- making specific queries using XNAT web services,
- data download conforming to specific naming convention and directory structure, using XNAT web services
- ensure all queries required to support processing workflow are working.
- Step 3: Data Processing
- Implement & execute the script-driven tractography workflow using web services,
- describe and upload results.
- ensure results are appropriately structured and named in repository, and queriable via web GUI and web services.
Participants
- sites involved: MGH NMR center, MGH Radiology, CHB Radiology
- number of users: ~10
- PI: Ellen Grant
- staff: Rudolph Pienaar
- clinicians
- IT staff
Outcome Metrics
Step 1: Data Management
- Visual confirmation (via web GUI) that all data is present, organized and named appropriately
- other?
Step 2: Query Formulation
- Successful tests that responses to XNAT queries for all MRIDs given a protocol name match results returned from currently-used search on the local filesystem.
- Query/Response should be efficient
Step 3: Data Processing
- Pipeline executes correctly
- Pipeline execution not substantially longer than when all data is housed locally
- other?
Overall
- Local disk space saved?
- Data management more efficient?
- Data management errors reduced?
- Barriers to sharing data lowered?
- Processing time reduced?
- User experience improved?
Fundamental Requirements
- System must be accessible 24/7
- System must be redundant (no data loss)
- Need a better client than current web GUI provides:
- faster
- PACS-like interface.
- image viewer should open in SAME window (not pop up a new)
- number of clicks to get to image view should be as few as possible.
Outstanding Questions
Plans for improving web GUI?
Data
Retrospective data consists of ~1787 studies, ~1TB total. Data consists of
- MR data, DICOM format
- Demographics from DICOM headers
- Subsequent processsing generates ".trk" files
- ascii text files ".txt"
- files that contain protocol information
Progress notes
Sprint 2
- Meeting on 4/08/2010: work has restarted on this effort. In the April/May timeframe:
- Wendy will revisit the tcl scripts and ensure that they are compliant with NEW XNAT 1.4 release.
- Rudolph and Dan Ginsburg will choose and provide three datasets for upload with these scripts.
- Wendy will upload these datasets using the previously developed mapping between Rudolph's data description and XNAT's schema.
- Wendy will send Dan a pointer to XNAT 1.4 documentation and tutorials.(Done: see here )
- After this upload, we will all visit and review organization of data on XNAT.
- Dan will also abstract their XML description of metadata used to populate their web-based front end, to make a friendlier adaptor for XNAT schema.
- Once approach for organizing and managing data on XNAT is settled, Wendy will begin writing scripts to
- make appropriate queries, and
- translate xml responses into appropriate format for web-based front end to display.
XNAT Evaluation - Dan G
- Installed XNAT 1.4 in Ubuntu 9.10 VM. Got tripped up a bit on issue that was solved on xnat_discussion list link
- Can't get user sessions to work, so I modified Wendy's scripts to pass -u/-p on each call, awaiting fix on xnat_discussion list link
- Next step is to go through xnat.xsd schema and see how it maps to the fields we are currently using in our own XML. Will post results on Wiki when done.
XNAT Schema Mapping - Dan G
I went through the exercise of looking at the data that we currently use in our custom XML to see if/where it could be found in the XNAT XML Schema. First, here is a sample entry from our XML:
<PatientRecord> <PatientID>...</PatientID> <Directory>...</Directory> <PatientName>....</PatientName> <PatientAge>...</PatientAge> <PatientSex>...</PatientSex> <PatientBirthday>...</PatientBirthday> <ImageScanDate>...</ImageScanDate> <ScannerManufacturer>...</ScannerManufacturer> <ScannerModel>...</ScannerModel> <SoftwareVer>...</SoftwareVer> <Scan>... </Scan> <Scan>... </Scan> <Scan>... </Scan> <Scan>.... </Scan> <User>...</User> <Group>...</Group> </PatientRecord>
The following table maps between each of these tags and the xnat.xsd schema in XNAT 1.4:
| Tag | XNAT Schema Equivalent |
|---|---|
| PatientID | xnat:subjectData/label (this is where Wendy's script stores the MRID), retrieved with (e.g., ./XNATRestClient -u USERNAME -p PASSWORD -host http://localhost:8080/xnat/ -m GET -remote /REST/projects/FNNDSC_test/subjects) |
| Directory | xnat:experiment/label |
| PatientName | xnat:imageSessionData/dcmPatientName |
| PatientAge | xnat:demographicData/xnat:abstractDemographicData/(dob,yob,age) |
| PatientSex | xnat:demographicData/gender |
| PatientBirthday | xnat:demographicData/xnat:abstractDemographicData/(dob,yob,age)
Q: I think dob/yob/age are mutually exclusive, so if age is put in I am not sure if DOB is available? |
| ImageScanDate | xnat:experimentData/date |
| ScannerManufacturer | xnat:imageSessionData/scanner/manufacturer
NOTE: It looks like there might be server config to have the drop-down have pre-defined values |
| ScannerModel | xnat:imageSessionData/scanner/model |
| SoftwareVer | There are a lot of useful fields in xnat:mrScanData, but I did not in particular find the software version.
Q: Should it be added? Did I miss it? |
| Scan | xnat:mrScanData/parameters/scanSequence |
| User, Group | I don't think these belong in here once we go to XNAT, we will have to re-evaluate how we do authentication. Ideally we should use the accounts in XNAT rather than what we do now. |
Sprint 1
- Trying experiment with Rudolph -- review of test data, and trial run of bulk upload script
- Wendy is working on pydicom script to optionally anonymize from this script
- and also working on pydicom script to parse headers and add "fileData" on upload
- also working on extending the XNATquery.tcl tool to perform flexible query/parse.
- Rudolph can have a look at the testdata upload on central & discuss with wendy
- if ok, rudolph can try upload script on central, or on chb instance (may be unexpected differences?)
Use Case Assessment #2
Helpful XNAT links
Use Case Assessment #1
Current Data Management Process
DICOM raw images are produced at radiology PACS at MGH, and are manually pushed to the PACS hosted on KAOS resided at MGH NMR center. The images are processed by a set of PERL scripts to be renamed and re-organized into a file structure where all images for a study are saved into a directory named for the study. DICOM images are currently viewed with Osiris on Macs.
Target Data Management Process (Step 1)
Step 1: Develop an Image Management System for BDC (IMS4BDC) with which at least the following can be done:
- Move images from MGH (KAOS) to a BDC machine at Children's
- Step 1a: Import legacy data into IMS4BDC from existing file structure and CDs
- Step 1b: Write scripts to execute upload of newly acquired data.
Target Query Formulation (Step 2)
Step 2. Develop Query capabilities using scripted client calls to XNAT web services, such as:
Show all subjectIDs scanned with protocol_name = ProtocolName Show all diffusion studies where patients ages are < 6
- Scripting capabilities: Scripts need to query and download data into appropriate directory structure, and support appropriate naming scheme to be compatible with existing processing workflow.
Target Processing Workflow (Step 3)
Step 3:
- Execute query/download scripts
- Run processing locally, on cluster, etc.
- Describe & upload processing results
- (eventually want to) Share images with clinical physicians
- (eventually want to) Export post-processed data back to clinical PACS
Fitting Data to XNAT Data Model
Test data from Rudolph
I think we have this mappint from project to XNE data model:
- MRID = SubjectID (1687 subjects?)
- each SubjectID may have single experiment, but multiple MRSessions within that experiment
- each "storage" directory for a particular MRID (in dcm_mrid.log) = MRSessionID
- each scan listed (int toc.txt) = ScanID in the MRsession
- important metadata contained in dicom headers, in dcm_mrid_age.log, dcm_mrid_age_days.log, and in the toc.txt file in each session directory.
This gives us a unique way to
- have unique subject IDs
- have unique MRSessionIDs for each subject,
- have unique scanIDs within each MRSession
- search for subject (by ID, age, or dicom header info) or
- search for image data by age (or dicom header info)
As regards anonymization
Rudolph doesn't specifically need XNAT to do the anonymization. Wants XNAT to contain all relevant data and where/if necessary export/transmit DICOM data anonymized.
Rudolph has own MatLAB script that can do batch anonymization -- but if possible XNAT package should probably provide a means for that.
Draft approach to uploading data for Rudolph
- Create a Project on the webGUI
- Write a webservices-client script that will batch:
- create subject (tested)
- create experiment for subject (tested)
- create mrsession for subject (tested)
- for each scan in mrsession
- anonymize (not sure)
- do dicom markup (not sure)
- add other metadata from toc.txt and *.log files (not sure)
- upload scan data into db (tested)
Current data (subset) organization
The top level directory contains
- a dcm_MRID.log file that contains a mapping between MRID's (PatientIDs?) and unique MRSessionNames
- a dcm_MRID_age.log file that maps MRID's to ages in months and years
- a dcm_MRID_age_days.log file tha tmaps MRID's to ages in days
- subdirectories named for MRSessions.
- each subdirectory contains a toc.txt file that includes patient and session information and a list of scans and scan types.
See examples below:
Questions sent to Rudolph about test data:
First, in the top-level dir, there are three log files: dcm_MRID.log dcm_MRID_age.log dcm_MRID_ageDays.log
1. do these files contain the MRIDs for *all* subjects in the entire retrospective study?
--Yes, at least current to the timestamp of the log file.
2. Some MRIDs appear to be purely numerical, and some alphanumerical. (3_S_658300). Is that correct?
Yes again. Unfortunately there seems to no standard technique for spec'ing the MRID number. This number, however, is the key most often used by clinicians, and thus is a primary key for the database. Problems abound, of course -- the MRID is linked to a single patient, but is not necessarily guaranteed to be unique. A patient keeps the same MRID, so multiple scans result in multiple instances. The combination of MRID+<storageDirectory> is unique (but also redundant, since the <storageDirectory> is unique, by definition. So essentially the log files are lookup tables for MRIDs and actual storage locations in the filesystem.
3. Two age files, one contains age in months or years (1687 entries) -- the other contains age in days (1687 entries):
- does this mean there are 1687 MRID's in total?
- what does age (days) = -1 mean?
These are just convenience files. Often times a typical 'query' would be: "findallMRID WHERE age IN <someAgeConstraint> AND protocol IN <someProtocolConstraint>."
The dcm_MRID_age.log maps the MRID to a storage location, and provides the age as tagged in the DICOM header. Of course, mixing different age formats (like 012M and 004W etc) isn't batch processing friendly. So, the dcm_MRID_ageDays.log converts all these age specifiers to days, and sorts the table on that field.
The '-1' means that some error occurred in the day calculation. Most likely, the associated age value wasn't present.
4. And the two dataset directories you shared:
Avanto-26039-20080130-134825-078000/
GENESIS_SIGNA-000000000000234-20041122-211850/
each directory contain data and a .toc file that includes:
- the "PatientID" is this equivalent to MRID?
- and some other info including age, scan date, etc.
- the filenames and scan types of a *set* of scans:
- collected in one MRsession on that scan date?
- or in the entire retrospective project?
- and is all the data for the set of scans listed contained in this directory?
true: PatientID == MRID The filenames and scantypes correspond to one session on that scan date. Other scan dates for that MRID will be in different directories. Essentially, the data is packed according to <scannerSpec>-<scanprefix>-<scandate>-<scantime>-<trailingID>.
Upload tests:
Remote script to upload -- parsing Rudolph's data and making webservices calls to create subjects:
Local data organization to XNAT data model mapping
implementing script to batch upload data from local to remote using webservices:
#----------------------------------------------------------------------------------------
# Local and Remote data organization notes
#----------------------------------------------------------------------------------------
# Batch upload of retrospective study maps local data organization
# to XNAT data model in the following way:
# xnat Project = PienaarGrant
# xnat Project contains list of unique xnat SubjectIDs (= local MRIDs)
# Each xnat SubjectID contains a list of xnat "Experiments"
# Each xnat Experiment is an "MRSession" with unique ID (and label = local dirname from dcm_mrid.txt)
# Each xnat MRSession labeled with local dirname contains set of xnat "Scans" (listed in local toc.txt)
# Each xnat Scan contains the image data.
# Project, Subjects, Experiments, and Scans can all have searchable metadata
# Projects, Experiments can have associated files (dcm_mrid.txt, toc.txt, error logs, etc.)
#----------------------------------------------------------------------------------------
# A schematic of the local organization looks like this:
#----------------------------------------------------------------------------------------
#
# Project root dir
# |
# { MRSession1, MRSession2, ..., MRSessionN} + {projectfiles = dcm_MRID.log, dcm_MRID_age.log, dcm_MRID_ageDays.log, error.log}
# |...
# { scanfiles=scan1.dcm, scan2.dcm,...,scanM.dcm } + {sessionfiles = toc.txt, toc.err, log/*, log_V/*}
#
#----------------------------------------------------------------------------------------
# A schematic of the xnat organization looks like this:
#----------------------------------------------------------------------------------------
#
# Project + {projectfiles.zip}
# |
# { SubjectID1, SubjectID2, ... SubjectIDN }
# ...|
# List of "Experiments" { MRSession, MRSession, ..., MRSession}
# ...|
# {scan1...scanN} + {sessionfiles.zip}
#
# An XNAT "Experiment" is an XNAT "imagingSession"
# An ImagingSession may be a MRSession, PETSession or CTSession (on central.xnat.org)
# -we are calling each local directory for a given MRID a new XNAT Experiment (MRSession).
# Each XNAT "imagingSession" contains a collection of XNAT "scans".
# we are calling each scan in a given local directory an XNAT "scan" in the MRSession.
Experiment to try with Rudolph
- Upload script is ready to be tested.
- Query and Download script still being developed
Resources needed to run the scripts
Scripts (ready to test for bulk upload)
Instructions & Notes
Rudolph:
Results of this script's upload are on central.xnat.org under the test project "PienaarGrantRetrospectiveTest". This is a private project because it contains protected medical information.
- once you make an account on central, send me your login and I'll add you to the list of users who can access the data.
- Then, click thru and let me know if the way data is organized appropriately matches your data.
- I've only uploaded the data for mrid=000000, and only one mrsession was in the test set -- so only one MRSession has scan data in it:
- GENESIS_SIGNA-000000000000234-20041122-211850
- if this doesn't work for you, let's meet and figure out how to fix.
- also, i've only tested for mrid=000000, and not for all mrids, so don't know whether there's a surprise when the code is run for all mrids in dcm_MRID.log.
- script is intended to work for initial bulk upload and for incremental uploads too (tho probably inefficiently) -- adding new experiments and their data as they appear in new dcm_MRID.log and other files. We'll have to test this.
to use:
- Create a test project using xnat web gui
- Using the "Manage" tab:
- Choose to make the project "private"
- Choose to place data directly into the databasee (not prearchive)
- Choose to skip the quarantine
- Download the XnatRESTClient (link above in 'resources' section)
- Download and install Tcl (link above in 'resources' section)
- Download the scripts and unzip (link above)
- Edit the XNATglobals.tcl file to customize
- run ./PGBulkUp.tcl -d rootDataDirectory
- the script will spew lots of text.
- some error messages pop up from time to time, and I'm not sure what causes the periodic croak:
- ...>>>>>>ERROR: AddScanData got unexpected response from remote PUT of scan data: The scan may not have been properly created. Perform QA check for: subject=000000 MRSession=CENTRAL_E00702 scan=Scan4 on remote host.
- ...>>>>>>ERROR: Got error response from XNAT host.
- ...>>>>>>ERROR: Got error message instead of experimentID from remote PUT of experiment: Experiment may not have been properly created. Please perform QA check for 000000 on remote host.
- So we should save the script output to see whether problems pop up.
PROJECT STATUS
- 9/29/09 - Scripts for bulk uploading using web services client made available for testing.
- Right now these scripts will upload entire retrospective study
- They do not parse DICOM headers and add header information to XNAT's "fileData"
- They do not anonymize
This seems like a reasonable first step since rudolph has his own scripts to anonymize, but did not want his data necessarily anonymized in the repository.