Difference between revisions of "2009 Summer Project Week MRSI-Module"
| Line 27: | Line 27: | ||
<div style="width: 30%; float: left;"> | <div style="width: 30%; float: left;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | We adapted the signal processing module to the 3T image data of the UIowa phantom data. | ||
| + | |||
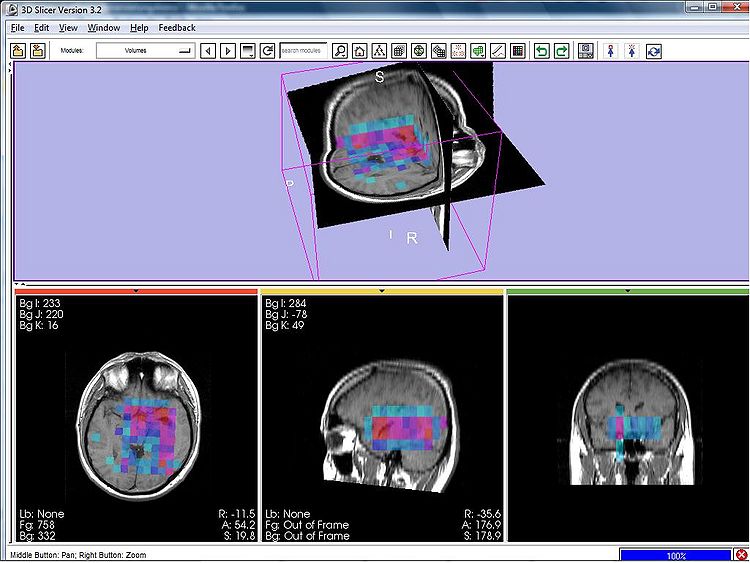
| + | We verified the registration of the spectroscopic images, now allowing to jointly visualize parametric maps (i.e., metabolic maps) of the MRSI signal processing module together with morphological images in Slicer (see Figure 1 below). | ||
| + | |||
| + | We defined an interface between the MRSI signal processing and the BrainCSI visualization module -- based on structured grids and later to be used in the 3DSlicer. | ||
| + | |||
</div> | </div> | ||
| + | |||
| + | |||
| + | [[File:Mrsi parametric-map-NAA 3d.jpg|thumpnail|750px]] | ||
| + | |||
| + | Figure 1 - 3D parametric map showing the NAA amplitudes of the spectra in a MR spectroscopic image in Slicer. | ||
Latest revision as of 14:49, 25 June 2009
Home < 2009 Summer Project Week MRSI-ModuleKey Investigators
- MIT: Bjoern Menze, Polina Golland
- Iowa: Jeff Yager, Vince Magnotta
Objective
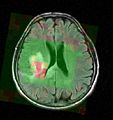
Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive diagnostic method used to determine the relative abundance of specific metabolites at arbitrary locations in vivo. Certain diseases -- such as tumors in brain, breast and prostate -- can be can be associated with characteristic changes in the metabolic level.
The objective of the current project is to develop a module proving the means for the processing and visualization of MRSI -- and thus for a joint analysis of magnetic resonance spectroscopic images together with other imaging modalities -- in Slicer.
Approach, Plan
Spectral fitting routines have been implemented, using a HSVD filter for water peak removal and baseline estimation, and a constrained non-linear least squares optimization for the fit of the resonance line models. MRSI post-processing routines for partial volume correction are available from the BrainCSI toolbox.
Our plan for the project week is to first integrate the spectral fitting routines into a Slicer module and to validate this module using MRSI phantom data. We then want to work towards interfacing signal processing module and post-processing tools.
Progress
We adapted the signal processing module to the 3T image data of the UIowa phantom data.
We verified the registration of the spectroscopic images, now allowing to jointly visualize parametric maps (i.e., metabolic maps) of the MRSI signal processing module together with morphological images in Slicer (see Figure 1 below).
We defined an interface between the MRSI signal processing and the BrainCSI visualization module -- based on structured grids and later to be used in the 3DSlicer.
Figure 1 - 3D parametric map showing the NAA amplitudes of the spectra in a MR spectroscopic image in Slicer.