Difference between revisions of "Slicer:Workshops:User Training 101"
From NAMIC Wiki
m (Update from Wiki) |
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
||
| (65 intermediate revisions by 11 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | [[Image:Nac.png|thumb|right|[http://nac.spl.harvard.edu Link to the '''NAC''' website]]] | |
| − | |||
| − | |||
= Introduction = | = Introduction = | ||
| + | This is a training compendium for using '''3D Slicer version 2'''. If you are looking for information about Slicer version 3, please follow this [https://www.slicer.org/wiki/Slicer3.6:Training#Software_tutorials Slicer3 101 page]. | ||
| − | + | This work is jointly sponsored by NA-MIC, NAC and NCIGT. | |
| − | + | This series of courses will teach you how to | |
| − | + | * load and view data | |
| − | * | + | * segment data and create 3D models |
| − | + | * perform Diffusion Tensor Imaging and fMRI analysis | |
| + | For an overview of Slicer, download this [[Media:NA-MIC-05-Slicer-Overview.ppt| slideshow]] | ||
| − | = Software | + | = Software = |
| + | <div class="floatleft"><span>[[Image:3DSlicer.png|80px|[[Image:3DSlicer.png| 3D Slicer Logo]]]]</span></div> | ||
| − | + | The current version of Slicer that is fully supported is Slicer 2.6. | |
| − | + | Please follow the [https://www.slicer.org/wiki/Slicer:Slicer2.6_Getting_Started Slicer 2.6 Getting Started] instructions to install the version of the Slicer program appropriate to your platform. | |
| − | = | + | = Training Compendium = |
| − | + | For course related questions, please send an e-mail to '''Sonia Pujol, Ph.D. (spujol at bwh.harvard.edu)'''. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
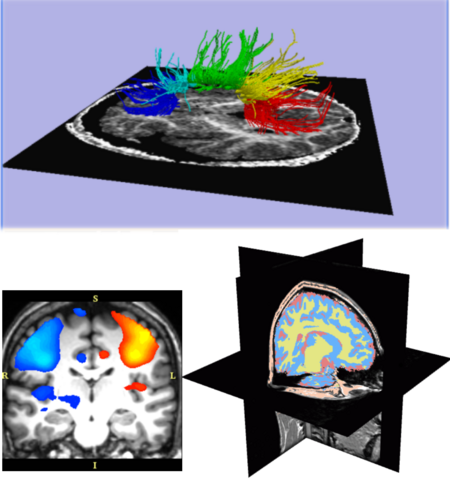
| − | + | [[Image:101.png|thumb|right|450px|'''Excerpts from the Slicer 101 courses:''' Diffusion Tensor Imaging fiber tractography (top); fMRI activation map (lower-left); automatic brain segmentation (lower-right).]] | |
| − | + | {| border="2" cellpadding="5" | |
| − | + | |- bgcolor="white" | |
| − | + | | | |
| − | + | | '''Course''' | |
| − | + | | '''Dataset''' | |
| − | + | |- bgcolor="white" | |
| − | + | ||[[Image:Training1 LoadingViewing.PNG|50px]] | |
| − | + | | | |
| − | + | [[Media:SlicerTraining1LoadingAndViewingData.ppt| Data Loading and Visualization ]] | |
| + | | | ||
| + | [[Media:Tutorial-with-dicom.zip|Tutorial_with_dicom.zip]] | ||
| + | |- bgcolor="white" | ||
| + | | [[Image:Training7Save.PNG|50px]] | ||
| + | | | ||
| + | [[Media:SlicerTraining7SavingData.ppt| Data Saving ]] | ||
| + | | | ||
| + | [[Media:Tutorial-with-dicom.zip|Tutorial-with-dicom.zip]] | ||
| + | |- bgcolor="white" | ||
| + | |[[Image:Training2Segmentation.PNG|50px]] | ||
| + | | | ||
| + | [[Media:Slicer_Segmentation_Tutorial.ppt| Manual Segmentation ]] | ||
| + | | | ||
| + | [[Media:Tutorial-with-dicom.zip|Tutorial_with_dicom.zip]] | ||
| + | |- bgcolor="white" | ||
| + | |[[Image:Training3LevelSets.PNG|50px]] | ||
| + | | | ||
| + | [[Media:03-LevelSet.ppt|Level-Set Segmentation ]] | ||
| + | | | ||
| + | [[Media:Tutorial-with-dicom.zip|Tutorial_with_dicom.zip]] | ||
| + | |- bgcolor="white" | ||
| + | | [[Image:Training10EM.png|50px]] | ||
| + | | | ||
| + | [[Media:SlicerAdvancedTraining_EMBrainAtlasClassifier_V1.0.ppt| Automatic Brain Segmentation ]] | ||
| + | | | ||
| + | [[Media:BrainAtlasClassifier.zip|BrainAtlasClassifier.zip ]] | ||
| + | |- bgcolor="white" | ||
| + | | [[Image:Training11Registration.PNG|55px]] | ||
| + | | | ||
| + | [[Media:SlicerAdvancedTraining11_Registration.ppt| Registration]] | ||
| + | | | ||
| + | [[Media:RegistrationSample.zip| RegistrationSample.zip]] | ||
| + | |- bgcolor="white" | ||
| + | |[[Image:Training4DTI.PNG|50px]] | ||
| + | | | ||
| + | [[Media:Slicer_DTMRI_Training4.ppt| Diffusion Tensor Imaging Analysis ]] | ||
| + | | | ||
| + | [[Media:SlicerSampleDTI.zip| SlicerSampleDTI.zip]] [[Media:Dwi-dicom.zip|Dwi-dicom.zip]] | ||
| + | |- bgcolor="white" | ||
| + | | [[Image:Training8Nrrd.PNG|50px]] | ||
| + | | | ||
| + | [[Media:SlicerTraining8-NrrdFileFormat.ppt| Nrrd File Format]] | ||
| + | | | ||
| + | [[Media:Tensor_data.zip|Tensor_data.zip]] | ||
| + | |- bgcolor="white" | ||
| + | | [[Image:Training9DicomToNrrd.PNG|50px]] | ||
| + | | | ||
| + | [[Media:SlicerTraining9_DTI-FromDicomToNrrd.ppt| Dicom to Nrrd Conversion ]] | ||
| + | | | ||
| + | [[Media:Dwi-dicom.zip|Dwi-dicom.zip]] | ||
| + | |- bgcolor="white" | ||
| + | | [[Image:Training5fMRI.PNG|50px]] | ||
| + | | | ||
| + | [[Media:SlicerTraining5_fMRI.ppt| Functional Magnetic Resonace Imaging Analysis ]] | ||
| + | | | ||
| + | [[Media:FMRIData1_short.zip|FMRIData1_short.zip]] [[Media:FMRIData2_long.zip|FMRIData2_long.zip]] | ||
| + | |- bgcolor="white" | ||
| + | | [[Image:Training6FreeSurfer.png|50px]] | ||
| + | | | ||
| + | [[Media:SlicerTraining6_vtkFreeSurferReaders.ppt| FreeSurfer Reader ]] | ||
| + | | | ||
| + | [[Media:FreeSurferSubjects.zip|FreeSurferSubjects.zip ]] | ||
| + | |} | ||
| − | = Slicer | + | = Slicer Additional Resources = |
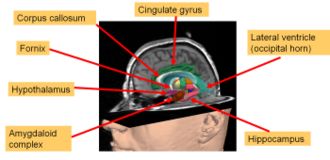
| + | [[Image:Limbic-2007.jpg|thumb|right|330px| A rendering of the Limbic System from the SPL PNL brain atlas]] | ||
| − | * * | + | * [[BrainAtlas|SPL PNL Brain Atlas]] |
| − | + | * [[Media:Training_EMLocalSegment_v1.pdf | EMSegmenter]] for multi-channel [[Media:EMSegmentTutorial.zip|data]] and white matter hyperintensities | |
| − | ** [[ | + | * [[DataFusion|Data Fusion and Registration]] |
| + | * [http://astromed.iic.harvard.edu/UsingSlicer Using 3DSlicer in Astronomy] | ||
| + | * [https://www.slicer.org/wiki/SlicerOnBIRN How to use Slicer with BIRN] | ||
| + | * CT based atlas of the [http://spl.harvard.edu/pages/Special:PubDB_View?dspaceid=1266 abdomen] for Slicer 3. The [[media:Abdominal_Atlas_Tutorial08.ppt|tutorial]] describing how to load and display the data in Slicer 3 | ||
| + | * [http://www.airwayinspector.org/ Airway inspector]: 3D Slicer based tool for morphometry analysis of the lung airways and emphysema. | ||
| − | = | + | = Feedback = |
| + | * [http://www.slicer.org/pages/Slicer_Community Feedback about Slicer] | ||
* [http://www.na-mic.org/Bug/ Bug reports] | * [http://www.na-mic.org/Bug/ Bug reports] | ||
| − | |||
= Acknowledgements = | = Acknowledgements = | ||
| + | *Slicer is being developed by a community of contributors. For more information see the [http://www.slicer.org Slicer] website. | ||
| − | + | <br /> Back to [[Training:Main|Training:Main]] | |
| − | + | [[Category:Slicer]] | |
Latest revision as of 18:07, 10 July 2017
Home < Slicer:Workshops:User Training 101Contents
Introduction
This is a training compendium for using 3D Slicer version 2. If you are looking for information about Slicer version 3, please follow this Slicer3 101 page.
This work is jointly sponsored by NA-MIC, NAC and NCIGT.
This series of courses will teach you how to
- load and view data
- segment data and create 3D models
- perform Diffusion Tensor Imaging and fMRI analysis
For an overview of Slicer, download this slideshow
Software
The current version of Slicer that is fully supported is Slicer 2.6.
Please follow the Slicer 2.6 Getting Started instructions to install the version of the Slicer program appropriate to your platform.
Training Compendium
For course related questions, please send an e-mail to Sonia Pujol, Ph.D. (spujol at bwh.harvard.edu).
| Course | Dataset | |
|
||
|
||
Slicer Additional Resources
- SPL PNL Brain Atlas
- EMSegmenter for multi-channel data and white matter hyperintensities
- Data Fusion and Registration
- Using 3DSlicer in Astronomy
- How to use Slicer with BIRN
- CT based atlas of the abdomen for Slicer 3. The tutorial describing how to load and display the data in Slicer 3
- Airway inspector: 3D Slicer based tool for morphometry analysis of the lung airways and emphysema.
Feedback
Acknowledgements
- Slicer is being developed by a community of contributors. For more information see the Slicer website.
Back to Training:Main