Difference between revisions of "Tissue Dependent Registration"
| (2 intermediate revisions by the same user not shown) | |||
| Line 28: | Line 28: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| + | Automatic segmentation into Skull, CSF, WM, GM of clinical dataset. | ||
| + | |||
Generate tissue dependent tetrahedral meshes. | Generate tissue dependent tetrahedral meshes. | ||
| Line 38: | Line 40: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | The Workshop allowed us to optimize the FE-computations to the level that we are able to generate enough samples to properly characterize the posterior on clinical datasets. | ||
| + | |||
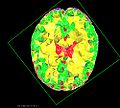
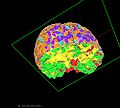
| + | The NAMIC kit is currently being utilized for visualizing results from the registration algorithm. Each MCMC sample provides a new configuration of the FE-mesh. We visualize this in realtime with each element colored according to the elastic parameter values. More interestingly, characterization of the posterior allows for quantification and visualization of | ||
| + | the uncertainty of the deformation field. We have developed different tools for visualizing and interacting with these uncertainty maps. | ||
| + | The work we performed at the project week has resulted in submission of a paper to Workshop on Biomedical Image Registration (WBIR) 2010 in Lubeck, Germany. | ||
</div> | </div> | ||
</div> | </div> | ||
<div style="width: 97%; float: left;"> | <div style="width: 97%; float: left;"> | ||
Latest revision as of 15:11, 8 February 2010
Home < Tissue Dependent RegistrationKey Investigators
- Petter Risholm (PhD Student, Harvard Medical School (BWH), University of Oslo)
- William Wells (Professor, Harvard Medical School (BWH) )
Objective
Most registration methods treat tissue parameters as uniform parameters over the image domain. For instance, when brain tissue is modeled as an elastic material, identical stiffness parameters are used for all tissue types (CSF, GM, WM). In our approach, we let each tissue type take on varying stiffness parameters and solve for the deformation as well as the stiffness parameters at the same time.
We pose the elastic registration problem as a Bayesian problem where we marginalize out the elastic parameters. A Markov Chain Monte Carlo (MCMC) sampling technique is applied to characterize the posterior.
We use a linear elastic Finite Element (FE) model to compute the likelihood (similarity term) and the deformation prior (regularizer). In a FE registration model, tissue is modeled by a mesh. To accurately model the tissue and let elastic parameters vary according to tissue type, we require a tissue dependent tetrahedral mesh. Assuming we have a segmentation of the brain tissue into CSF, GM and WM, we need a mesh where the tetrahedra conform to the different tissue types (the edges of a tetrahedra do not cross from one tissue type to the other but are either fully within a tissue type or on the boundary between two tissue types).
To properly characterize the posterior with a MCMC approach, a large number of samples is often needed. The FE-computations are the bottleneck as the FE-system has to be constructed and computed for each new sample generated. Hence, it is necessary with fast FE computations to generate enough samples.
Approach, Plan
Automatic segmentation into Skull, CSF, WM, GM of clinical dataset.
Generate tissue dependent tetrahedral meshes.
Optimize FE-computations.
Develop tool for visualizing a tetrahedral mesh with varying elastic parameters.
Progress
The Workshop allowed us to optimize the FE-computations to the level that we are able to generate enough samples to properly characterize the posterior on clinical datasets.
The NAMIC kit is currently being utilized for visualizing results from the registration algorithm. Each MCMC sample provides a new configuration of the FE-mesh. We visualize this in realtime with each element colored according to the elastic parameter values. More interestingly, characterization of the posterior allows for quantification and visualization of the uncertainty of the deformation field. We have developed different tools for visualizing and interacting with these uncertainty maps.
The work we performed at the project week has resulted in submission of a paper to Workshop on Biomedical Image Registration (WBIR) 2010 in Lubeck, Germany.