Difference between revisions of "Projects:RuleBasedStriatumSegmentation"
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|NA-MIC Collaborations]], [[Algorithm: | + | Back to [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|NA-MIC Collaborations]], [[Algorithm:Stony Brook|Stony Brook University Algorithms]], [[DBP1:Harvard|Harvard DBP1]], [[Engineering:Kitware|Kitware Engineering]], [[Engineering:Isomics|Isomics Engineering]], [[DBP1:Irvine|Irvine DBP1]] |
__NOTOC__ | __NOTOC__ | ||
= Rule Based Striatum Segmentation = | = Rule Based Striatum Segmentation = | ||
Latest revision as of 00:59, 16 November 2013
Home < Projects:RuleBasedStriatumSegmentationBack to NA-MIC Collaborations, Stony Brook University Algorithms, Harvard DBP1, Kitware Engineering, Isomics Engineering, Irvine DBP1
Rule Based Striatum Segmentation
In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules. We have implemented our code in Slicer 2. We currently provide modules for the semi-automatic segmentation of the DLPFC and the Striatum.
Description
We have developed an algorithm for Semi-Automatic Segmentation of the DLPFC based on the rules of Core 3 collaborator, Dr. James Fallon. This algorithm was tested last year in Matlab with successful results. This year, we implemented the algorithm into a 3D SLICER module which works with the current Editor Tab. A screenshot of the module is shown below. The ITK Bayesian Segmentation Filter is currently being incorporated into the module. This is important, since we use Bayesian classifiers in order to enhance the Fallon method. The motivation of the DLPFC semi-automatic segmenter was to minimize segmentation time of the DLPFC by incorporating the rules of Dr. Fallon into an algorithm, while still giving the user control of the segmentation process. The time to segment the DLPFC was reduced from over 30 minutes to approximately 5 minutes. The algorithm is based on the average proportional distances of the posterior boundary from the temporal lobe tip and the anterior boundary from the frontal pole. Each hemisphere must be done separately. The average shape is a parallelogram from the movement of the middle frontal gyrus dorsally as moving posteriorly through coronal slices Dr. James Fallon has visited Georgia Tech in December 2005 to train our local researchers about his heuristic rules. He will be visiting again on May 17-18, 2007 for further testing and algorithmic development as well as clinical applications.
Striatum Progress
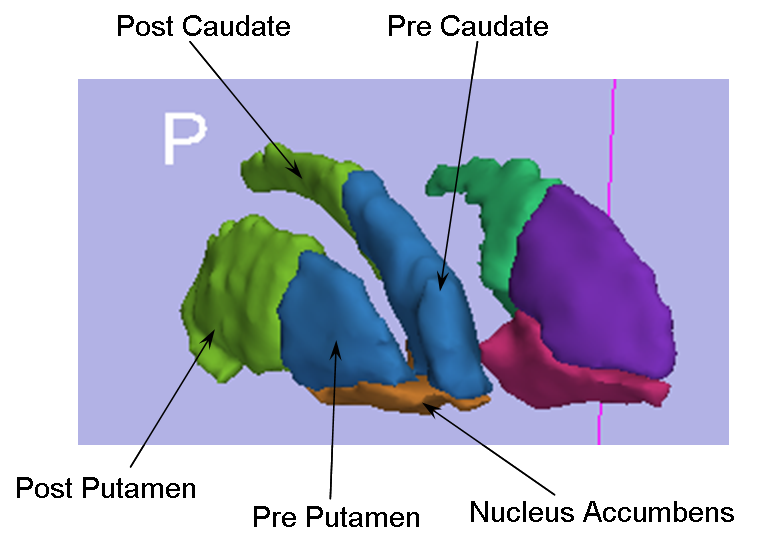
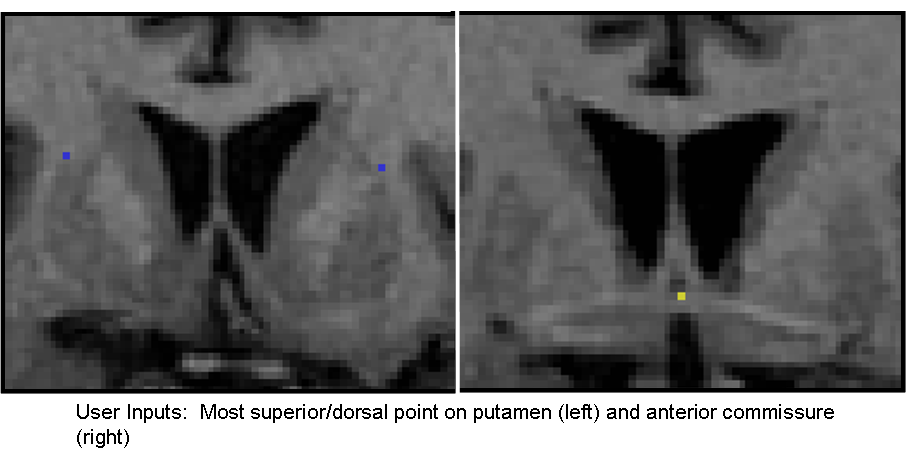
We have developed an algorithm for delineation of the striatum into 5 physiological subregions (pre/post caudate, pre/post putamen, and nucleus accumbens) while requiring only minimal user input. We have implemented this algorithm from the geometric rules for delineating the striatum as defined by our Core 3 collaborator, Dr. James Levitt of the PNL, into a 3D SLICER module. The current run time for the algorithm is ~20 seconds after the initial user input. The user inputs a label map of the full striatum, the most superior/dorsal voxel of the putamen on each slice, and the anterior commisure voxel (see figure below). From these, the labelmap is delineated into the aforementioned subregions. The figure below shows a 3D model of the left and right striatum delineated into the five subregions.
Striatum Representative Image and Descriptive Caption
Key Investigators
- Georgia Tech Algorithms: Ramsey Al-Hakim, Delphine Nain, Allen Tannenbaum, John Melonakos
- Harvard DBP1: Sylvain Bouix, James Levitt, Marc Niethammer, Martha Shenton.
- Kitware Engineering: Luis Ibanez
- Isomics Engineering: Steve Pieper
- Irvine DBP1: James Fallon
Publications
In Print