Difference between revisions of "2011 Winter Project Week:NerveSeg"
From NAMIC Wiki
| (5 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
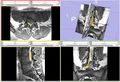
Image:NergeSeg-scr1.png|Figure 1 - Example of Manual Nerve Segmentation | Image:NergeSeg-scr1.png|Figure 1 - Example of Manual Nerve Segmentation | ||
Image:NerveSeg-scr2.png|Figure 2 - Example of results so far. | Image:NerveSeg-scr2.png|Figure 2 - Example of results so far. | ||
| + | Image:NerveSeg-scr3.png|Figure 3 - Example of results so far (slicer). | ||
| + | |||
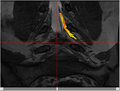
| + | Image:NerveSeg-res-scr1.png|Figure 4 - Example of automatic segmentation on Thursday afternoon for multiple nerves. | ||
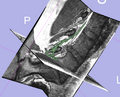
| + | Image:NerveSeg-res-scr2.png|Figure 5 - Complete render of nerve segmentations. | ||
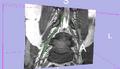
| + | Image:NerveSeg-res-scr3.png|Figure 6 - Another View. | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
* MIT: Adrian Dalca, Polina Golland | * MIT: Adrian Dalca, Polina Golland | ||
| − | * BWH | + | * BWH: Giovanna Danagoulian, Ehud Schmidt |
| + | * SPL: Ron Kikinis | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 28: | Line 34: | ||
# Select overall tracks at the end of the algorithm | # Select overall tracks at the end of the algorithm | ||
# Clean tracks from unimportant branches. | # Clean tracks from unimportant branches. | ||
| + | |||
| + | We hope to achieve #1 in the project week. | ||
</div> | </div> | ||
| Line 33: | Line 41: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | --- | + | * Received many ideas on new approaches. Thank you to everyone. |
| + | * Tried center-line search to avoid direct segmentation. Level-set or similar segmentation might follow. | ||
| + | ** Method is very sensitive to spilling into surrounding tissue. | ||
| + | ** Might be able to clean up bad centerlines with global nerve measure. | ||
| + | * Method of gradients (and intensity together) with other constraints (mostly intensity ranges) seems to be best right now after some development. | ||
| + | ** See Figures 4-6 | ||
| + | ** But still problematic in more ambiguous regions (neighboring anatomy of similar intensity) | ||
| + | *** Hopefully fix this with global measure. | ||
| Line 41: | Line 56: | ||
<div style="width: 97%; float: left;"> | <div style="width: 97%; float: left;"> | ||
| − | + | ||
</div> | </div> | ||
Latest revision as of 21:10, 13 January 2011
Home < 2011 Winter Project Week:NerveSegKey Investigators
- MIT: Adrian Dalca, Polina Golland
- BWH: Giovanna Danagoulian, Ehud Schmidt
- SPL: Ron Kikinis
Objective
We are developing a nerve segmentation algorithm for the automatic isolation of nerves and nerve ganglia inside the spinal sack and out through the vertebrae in new MR Myelography images.
Approach, Plan
Currently we use a particle-filter tracking approach for segmenting the nerves. The algorithm is given a seed point, preferably somewhere in the spine. The particles are tubes following Bézier curves (and hence forming a B-spline track). To compute the likelihood of each sample we are experimenting with measures that use image intensity and gradients within the patch. Correct partial volume estimates are important in the measure as well.
Current results are promising, but several parts have to be perfected:
- Finalize the measure - currently the best results are only using gradient values, we need to incorporate image intensity better
- Select overall tracks at the end of the algorithm
- Clean tracks from unimportant branches.
We hope to achieve #1 in the project week.
Progress
- Received many ideas on new approaches. Thank you to everyone.
- Tried center-line search to avoid direct segmentation. Level-set or similar segmentation might follow.
- Method is very sensitive to spilling into surrounding tissue.
- Might be able to clean up bad centerlines with global nerve measure.
- Method of gradients (and intensity together) with other constraints (mostly intensity ranges) seems to be best right now after some development.
- See Figures 4-6
- But still problematic in more ambiguous regions (neighboring anatomy of similar intensity)
- Hopefully fix this with global measure.