Difference between revisions of "2011 Winter Project Week:Intra-ProceduralProstateMotion"
(→Data) |
|||
| (24 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | <gallery> | + | <gallery perrow=5> |
| − | Image:PW- | + | Image:PW-SLC2011.png|[[2011_Winter_Project_Week#Projects|Projects List]] |
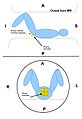
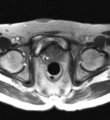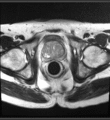
Image:Transperineal_prostate_biopsy.jpg|Patient configuration for transperineal prostate biopsy in closed bore MRI | Image:Transperineal_prostate_biopsy.jpg|Patient configuration for transperineal prostate biopsy in closed bore MRI | ||
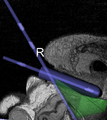
Image:Transrectal_prostate_biopsy.jpg|Patient configuration for transrectal prostate biopsy in closed bore MRI | Image:Transrectal_prostate_biopsy.jpg|Patient configuration for transrectal prostate biopsy in closed bore MRI | ||
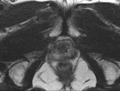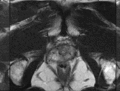
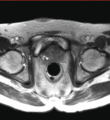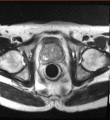
Image:Pelvis_and_prostate_motion_example.gif|Motion of the pelvis and prostate gland over the first 30 minutes of MR-guided transperineal biopsy procedure. | Image:Pelvis_and_prostate_motion_example.gif|Motion of the pelvis and prostate gland over the first 30 minutes of MR-guided transperineal biopsy procedure. | ||
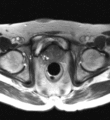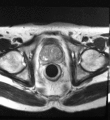
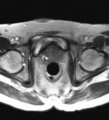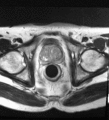
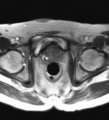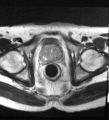
Image:TransrectalBiopsySagittal.png|Sagittal view of a transrectal MR-guided transrectal biopsy procedure. | Image:TransrectalBiopsySagittal.png|Sagittal view of a transrectal MR-guided transrectal biopsy procedure. | ||
| + | Image:prostate_unreg.gif|Unregistered | ||
| + | Image:prostate_rega.gif|Registered w/ axial slice | ||
| + | Image:prostate_regac.gif|Registered w/ axial+cor slices | ||
| + | Image:prostate_regacs.gif|Registered w/ axial+cor+sag slices | ||
| + | Image:prostate_regvol.gif|Registered w/ full volume | ||
</gallery> | </gallery> | ||
| Line 31: | Line 36: | ||
* discuss with Slicer engineering core plans and challenges in improving support for deformable transformations in 3D Slcer | * discuss with Slicer engineering core plans and challenges in improving support for deformable transformations in 3D Slcer | ||
| − | Experimental setup: | + | '''Experimental setup''': compare performance of the sparse volume registration tools, and correlate with the full volume registration results; this experiment will be done (1) between the volume obtained in the beginning of the procedure and needle conf volume and (2) between two needle confirmation volumes both for TR and TP Bx approaches for two patients. |
| − | + | * Experiment 1: full volume registration | |
| − | * Experiment 1: full volume registration | + | * Experiment 2: sparse volume registration by masking individual slices in the full volume: Axial slice, Axial+Cor slices, Axial+Cor+Sag slices |
| − | * Experiment 2: sparse volume registration by masking individual slices in the full volume: | ||
| − | |||
* Comparison metrics: difference wrt the transform obtained in Exp. 1, execution time | * Comparison metrics: difference wrt the transform obtained in Exp. 1, execution time | ||
</div> | </div> | ||
| Line 47: | Line 50: | ||
<h3>Dissemination</h3> | <h3>Dissemination</h3> | ||
| − | http://viewvc.slicer.org/viewcvs.cgi/trunk/ | + | http://viewvc.slicer.org/viewcvs.cgi/trunk/IGTLoadableModules/ProstateRegistration/?root=NAMICSandBox |
| − | |||
| − | |||
| − | |||
==Data== | ==Data== | ||
| Line 70: | Line 70: | ||
*TP11: Case011 | *TP11: Case011 | ||
| − | == | + | ==Found bugs== |
*itkOptImageToImageMetric gives different result if executed on single vs. multiple threads. | *itkOptImageToImageMetric gives different result if executed on single vs. multiple threads. | ||
| − | *ImageSpatialObject< TDimension, PixelType >::ValueAt evaluates the value at a different position than ImageMaskSpatialObject< TDimension >::IsInside (which determines if the point is inside or not) | + | *ImageSpatialObject< TDimension, PixelType >::ValueAt evaluates the value at a different position than ImageMaskSpatialObject< TDimension >::IsInside (which determines if the point is inside or not) => [http://public.kitware.com/Bug/view.php?id=11699 0011699: ImageMaskSpatialObject IsInside and ValueAt methods give inconsistent results] |
| + | |||
| + | ==BWH slice-volume rigid registration== | ||
| + | * VolumeToSliceRegistration (BWH): rigidly register initial multi-slice intra-procedural volume to the needle confirmation slice/volume. Needle confirmation volume is fixed. Gaussian smoothing is applied to needle confirmation scan. Metric is NMI, optimizer is VersorRigid3DTransformOptimizer. The code accepts binary mask that allows to restrict the metric sampling region in the fixed volume. Source code: http://viewvc.slicer.org/viewcvs.cgi/trunk/SLC2011-ProstateRegistration/SliceToVolumeRegistration/SliceRegistration.cxx | ||
==Queen's registration algo steps:== | ==Queen's registration algo steps:== | ||
| − | * | + | *Rescale the fixed image to have the same min/max intensity as the moving image |
| − | * | + | *Set up fixed image regions: VOI is only the prostate |
| − | + | *Set up all supported metrics: MeanSquaresSparseVolumeToVolumeMetric + NC2SparseVolumeToVolumeMetric + ViolaWellsSparseVolumeToVolumeMetric | |
| − | * | + | *Set multiple fixed image regions (one for each slice) for all the metrics |
| − | * | + | *Register: CMAEvolutionStrategyOptimizer, rigid translation, mutual information metric; CenteredTransformInitializer; both volumes smoothed |
| − | * | + | *Apply determined transform |
| − | * | + | *Register: RegularStepGradientDescentOptimizer (constrain translation), Euler 3D transform, mutual information metric; CenteredTransformInitializer; both volumes smoothed |
| − | * | + | *Apply determined transform |
| − | * | + | *Register: MeanSquaresSparseVolumeToVolumeMetric/NC2SparseVolumeToVolumeMetric/ViolaWellsSparseVolumeToVolumeMetric, RegularStepGradientDescentOptimizer, affine transform, |
| − | * | + | *Apply determined transform |
| − | * | + | *Register: bspline transform 5 nodes, same metric, GD optimizer |
| − | * | + | *Register: bspline transform 15 nodes, same metric, GD optimizer |
| − | * | + | |
| + | ==Results== | ||
| + | |||
| + | Reported values are average (maximum) difference in mm over the volume of interest (prostate gland) between the transform obtained using whole volume needle confirmation image VS sparse volume needle confirmation images (axial, axial+cor, axial+cor+sag slices). | ||
| + | |||
| + | {| border="1" cellpadding="5" cellspacing="0" align="center" | ||
| + | |- | ||
| + | | align="left" style="background:#e0e0e0;" | '''High-res volume''' | ||
| + | | align="center" style="background:#e0e0e0;" colspan=4 | '''Needle confirmation volume 1''' | ||
| + | | align="center" style="background:#e0e0e0;" colspan=4 | '''Needle confirmation volume 2''' | ||
| + | |- | ||
| + | | align="left" | | ||
| + | | align="left" | '''Mis-registration magnitude recovered by volume-to-volume registration''' | ||
| + | | align="center | '''Axial slice''' | ||
| + | | align="center" | '''Axial+Cor slice''' | ||
| + | | align="center" | '''Axial+Cor+Sag slice''' | ||
| + | | align="left" | '''Mis-registration magnitude recovered by volume-to-volume registration''' | ||
| + | | align="center" | '''Axial slice''' | ||
| + | | align="center" | '''Axial+Cor slice''' | ||
| + | | align="center" | '''Axial+Cor+Sag slice''' | ||
| + | |- | ||
| + | | align="left" | '''TR03''' | ||
| + | | align="left" | '''1.8 (3.3)''' bone reg. error <1mm | ||
| + | | align="left" | 0.9 (2.2) | ||
| + | | align="left" | 0.9 (2.1) | ||
| + | | align="left" | 0.8 (1.8) | ||
| + | | align="left" | '''2.0 (3.0)''' bone reg. error <1mm | ||
| + | | align="left" | 0.9 (2.1) | ||
| + | | align="left" | 0.9 (2.1) | ||
| + | | align="left" | 0.8 (1.9) | ||
| + | |- | ||
| + | | align="left" | '''TR02''' | ||
| + | | align="left" | '''7.4 (12.1)''' bone reg. error <1mm | ||
| + | | align="left" | 3.1 (6.7) | ||
| + | | align="left" | 2.8 (6.3) | ||
| + | | align="left" | 2.6 (6.1) | ||
| + | | align="left" | '''6.7 (10.7)''' bone reg. error <1mm | ||
| + | | align="left" | 4.1 (9.4) | ||
| + | | align="left" | 3.9 (8.8) | ||
| + | | align="left" | 4.0 (9.1) | ||
| + | |- | ||
| + | | align="left" | '''TR01''' | ||
| + | | align="left" | '''2.3 (3.1)''' bone reg. error <1mm (small image) | ||
| + | | align="left" | 1.2 (2.3) | ||
| + | | align="left" | 1.2 (2.3) | ||
| + | | align="left" | 1.3 (2.3) | ||
| + | | align="left" | '''2.5 (3.0)''' bone reg. error <2mm (small image) | ||
| + | | align="left" | 1.5 (3.3) | ||
| + | | align="left" | 1.5 (3.3) | ||
| + | | align="left" | 1.6 (3.5) | ||
| + | |- | ||
| + | | align="left" | '''TP9''' | ||
| + | | align="left" | '''5.2 (7.1)''' bone reg. error <1mm (small image) | ||
| + | | align="left" | 0.9 (2.0) | ||
| + | | align="left" | 0.8 (1.8) | ||
| + | | align="left" | 0.6 (1.3) | ||
| + | | align="left" | na | ||
| + | | align="left" | na | ||
| + | | align="left" | na | ||
| + | | align="left" | na | ||
| + | |- | ||
| + | | align="left" | '''TP10''' | ||
| + | | align="left" | '''4.5 (6.7)''' bone reg. error <1mm | ||
| + | | align="left" | 1.7 (2.0) | ||
| + | | align="left" | 1.7 (1.9) | ||
| + | | align="left" | 1.5 (2.1) | ||
| + | | align="left" | '''1.2 (1.4)''' bone reg. error <1mm | ||
| + | | align="left" | 0.3 (0.7) | ||
| + | | align="left" | 0.3 (0.5) | ||
| + | | align="left" | 0.3 (0.5) | ||
| + | |- | ||
| + | | align="left" | '''TP11''' | ||
| + | | align="left" | '''1.9 (2.5)''' bone reg. error <1mm | ||
| + | | align="left" | 0.3 (0.5) | ||
| + | | align="left" | 0.2 (0.4) | ||
| + | | align="left" | 0.2 (0.4) | ||
| + | | align="left" | '''2.2 (2.7)''' bone reg. error <1mm | ||
| + | | align="left" | 0.3 (0.6) | ||
| + | | align="left" | 0.2 (0.6) | ||
| + | | align="left" | 0.2 (0.6) | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Observations: | ||
| + | *Volume-to-volume registration of intra-procedural hi-resolution initial image and needle confirmation image registers the bones quite accurately, thus efficiently compensates gross patient motion (checked by looking at the registered volumes, error seem to be <1mm) | ||
| + | *Slice-to-volume registration compensates large part of the patient motion (by about 50-80%) | ||
| + | *The registration doesn't compensate prostate motion. To compensate that the registration a second registration step should be performed, that uses a smaller ROI (containing only the prostate). A third step could be to perform non-rigid registration of the prostate. | ||
==References== | ==References== | ||
Latest revision as of 00:15, 25 January 2011
Home < 2011 Winter Project Week:Intra-ProceduralProstateMotionKey Investigators
- BWH: Andrey Fedorov
- Queens University: Andras Lasso
Objective
Multi-parametric diagnostic MRI was shown to be important in localization of prostate cancer. MR-guided prostate cancer biopsy (using transperineal access at BWH, transrectal access at Queen's clinical partners) performed in the closed bore MR scanner relies on image registration between the intra-procedural configuration of the prostate gland and the pre-procedural diagnostic MRI to locate the suspected cancer regions and guide the biopsy sample collection. Intra-operative image acquisition includes a volumetric scan of the whole prostate gland in the beginning of the procedure, followed by single-slice (or few orthogonal slices) acquisitions throughout the procedure with the purpose of confirming biopsy needle location. In the current image processing workflow, the diagnostic MRI is registered with the initial intra-procedural MRI.
One of the challenges we are facing is the motion and deformation of the prostate gland during the course of the procedure, which is typically ~1 hour long. Significant patient motion may invalidate the result of the initial pre- to intra-procedural MRI registration.
Our objective is to develop robust image acquisition and registration protocols to detect and compensate for the intra-procedural prostate motion/deformation, preferably using single-slice acquisitions throughout the procedure.
Approach, Plan
- evaluate and compare the solutions developed at BWH and Queens for volume-to-slice registration on the common data
- seek input from NA-MIC registration experts on the design of the registration approach
- discuss the high-level approach and plans for integrating volume-to-slice registration functionality into ProstateNav Slicer module
- discuss with Slicer engineering core plans and challenges in improving support for deformable transformations in 3D Slcer
Experimental setup: compare performance of the sparse volume registration tools, and correlate with the full volume registration results; this experiment will be done (1) between the volume obtained in the beginning of the procedure and needle conf volume and (2) between two needle confirmation volumes both for TR and TP Bx approaches for two patients.
- Experiment 1: full volume registration
- Experiment 2: sparse volume registration by masking individual slices in the full volume: Axial slice, Axial+Cor slices, Axial+Cor+Sag slices
- Comparison metrics: difference wrt the transform obtained in Exp. 1, execution time
Dissemination
Data
- TR01: 20101021_PMH
- Not much patient motion (about 1-2mm)
- TR02: 20100721_JHU
- Large (abour 3-5mm) patient motion between T2 and needle confirmation image
- Small patient motion between needle confirmation images
- Registration doesn't work with 1 slice (it gives identity matrix as a result): File: c:\users\andras\devel\slicer-3.6\slicer3-lib\insight\code\review\itkOptMattesMutualInformationImageToImageMetric.txx. Line: 1046. Description: itk::ERROR: MattesMutualInformationImageToImageMetric(01012928): Joint PDF summed to zero
- Registration does work with 3 slices
- TR03: 20101216_NIH
- Slight rotation between T2 and needle confirmation image
- Negligible patient motion between needle confirmation images
- DoRegistration doesn't work with 1 slice (it gives identity matrix as a result): Optimizer stop condition: VersorRigid3DTransformOptimizer: Gradient magnitude tolerance met after 0 iterations. Gradient magnitude (0) is less than gradient magnitude tolerance (0.0001).
- Registration does work with 3 slices
- TP09: Case009
- TP10: Case010
- TP11: Case011
Found bugs
- itkOptImageToImageMetric gives different result if executed on single vs. multiple threads.
- ImageSpatialObject< TDimension, PixelType >::ValueAt evaluates the value at a different position than ImageMaskSpatialObject< TDimension >::IsInside (which determines if the point is inside or not) => 0011699: ImageMaskSpatialObject IsInside and ValueAt methods give inconsistent results
BWH slice-volume rigid registration
- VolumeToSliceRegistration (BWH): rigidly register initial multi-slice intra-procedural volume to the needle confirmation slice/volume. Needle confirmation volume is fixed. Gaussian smoothing is applied to needle confirmation scan. Metric is NMI, optimizer is VersorRigid3DTransformOptimizer. The code accepts binary mask that allows to restrict the metric sampling region in the fixed volume. Source code: http://viewvc.slicer.org/viewcvs.cgi/trunk/SLC2011-ProstateRegistration/SliceToVolumeRegistration/SliceRegistration.cxx
Queen's registration algo steps:
- Rescale the fixed image to have the same min/max intensity as the moving image
- Set up fixed image regions: VOI is only the prostate
- Set up all supported metrics: MeanSquaresSparseVolumeToVolumeMetric + NC2SparseVolumeToVolumeMetric + ViolaWellsSparseVolumeToVolumeMetric
- Set multiple fixed image regions (one for each slice) for all the metrics
- Register: CMAEvolutionStrategyOptimizer, rigid translation, mutual information metric; CenteredTransformInitializer; both volumes smoothed
- Apply determined transform
- Register: RegularStepGradientDescentOptimizer (constrain translation), Euler 3D transform, mutual information metric; CenteredTransformInitializer; both volumes smoothed
- Apply determined transform
- Register: MeanSquaresSparseVolumeToVolumeMetric/NC2SparseVolumeToVolumeMetric/ViolaWellsSparseVolumeToVolumeMetric, RegularStepGradientDescentOptimizer, affine transform,
- Apply determined transform
- Register: bspline transform 5 nodes, same metric, GD optimizer
- Register: bspline transform 15 nodes, same metric, GD optimizer
Results
Reported values are average (maximum) difference in mm over the volume of interest (prostate gland) between the transform obtained using whole volume needle confirmation image VS sparse volume needle confirmation images (axial, axial+cor, axial+cor+sag slices).
| High-res volume | Needle confirmation volume 1 | Needle confirmation volume 2 | ||||||
| Mis-registration magnitude recovered by volume-to-volume registration | Axial slice | Axial+Cor slice | Axial+Cor+Sag slice | Mis-registration magnitude recovered by volume-to-volume registration | Axial slice | Axial+Cor slice | Axial+Cor+Sag slice | |
| TR03 | 1.8 (3.3) bone reg. error <1mm | 0.9 (2.2) | 0.9 (2.1) | 0.8 (1.8) | 2.0 (3.0) bone reg. error <1mm | 0.9 (2.1) | 0.9 (2.1) | 0.8 (1.9) |
| TR02 | 7.4 (12.1) bone reg. error <1mm | 3.1 (6.7) | 2.8 (6.3) | 2.6 (6.1) | 6.7 (10.7) bone reg. error <1mm | 4.1 (9.4) | 3.9 (8.8) | 4.0 (9.1) |
| TR01 | 2.3 (3.1) bone reg. error <1mm (small image) | 1.2 (2.3) | 1.2 (2.3) | 1.3 (2.3) | 2.5 (3.0) bone reg. error <2mm (small image) | 1.5 (3.3) | 1.5 (3.3) | 1.6 (3.5) |
| TP9 | 5.2 (7.1) bone reg. error <1mm (small image) | 0.9 (2.0) | 0.8 (1.8) | 0.6 (1.3) | na | na | na | na |
| TP10 | 4.5 (6.7) bone reg. error <1mm | 1.7 (2.0) | 1.7 (1.9) | 1.5 (2.1) | 1.2 (1.4) bone reg. error <1mm | 0.3 (0.7) | 0.3 (0.5) | 0.3 (0.5) |
| TP11 | 1.9 (2.5) bone reg. error <1mm | 0.3 (0.5) | 0.2 (0.4) | 0.2 (0.4) | 2.2 (2.7) bone reg. error <1mm | 0.3 (0.6) | 0.2 (0.6) | 0.2 (0.6) |
Observations:
- Volume-to-volume registration of intra-procedural hi-resolution initial image and needle confirmation image registers the bones quite accurately, thus efficiently compensates gross patient motion (checked by looking at the registered volumes, error seem to be <1mm)
- Slice-to-volume registration compensates large part of the patient motion (by about 50-80%)
- The registration doesn't compensate prostate motion. To compensate that the registration a second registration step should be performed, that uses a smaller ROI (containing only the prostate). A third step could be to perform non-rigid registration of the prostate.
References
- H. Tadayyon, A. Lasso, S. Gill, A. Kaushal, P. Guion, and G. Fichtinger, Target Motion Compensation in MRI-guided Prostate Biopsy with Static Images, EMBC2010 - 32nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 2010
- H. Tadayyon, MRI-Guided Prostate Motion Tracking using Multislice-to-Volume Registration, MASc Thesis, 2010