Difference between revisions of "2011 Summer Project Week White Matter Laterality"
From NAMIC Wiki
(Created page with '__NOTOC__ <gallery> Image:PW-MIT2011.png|Projects List Image:notfound.png|Interesting picture to be added... </gallery> '''Full Title of P…') |
|||
| (13 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2011.png|[[2011_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2011.png|[[2011_Summer_Project_Week#Projects|Projects List]] | ||
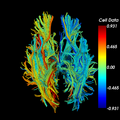
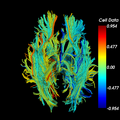
| − | Image: | + | Image:fiberLI-beforemidsagalign.png|before automatic midsagittal alignment |
| + | Image:fiberLI-aftermidsagalign.png|after automatic midsagittal alignment. Note increased symmetry (green laterality indices, near 0) | ||
</gallery> | </gallery> | ||
| − | ''' | + | '''White Matter Laterality/Asymmetry Measurement''' |
==Key Investigators== | ==Key Investigators== | ||
| − | * Harvard Medical School: | + | * Harvard Medical School: Lauren O'Donnell |
| − | + | * Kitware: JC (Thanks for help on how to integrate as a Slicer4 module/extension) | |
| + | * Harvard Medical School: Sandy Wells (Thanks for helpful discussions on angle representation and objective functions for registration) | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 16: | Line 18: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | #: | + | #:Fast computation of white matter |
| − | #: | + | #:fiber tract laterality indices |
| − | #: | + | #: in python and ... Slicer4 |
</div> | </div> | ||
| Line 25: | Line 27: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | #: | + | #:So far have preliminary code running in python/vtk |
| − | #:for | + | #:Slicer-ification is goal for this week |
| − | #: | + | #:Another goal: learn how to make an extension module |
| + | #:Another goal: can this module use testing framework | ||
</div> | </div> | ||
| Line 34: | Line 37: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | #: Project is now organized as a python package | ||
| + | #: Command line script written to run on large groups | ||
| + | #: Initial testing on R/L handedness group data showed significant differences | ||
| + | #: Preprocessing and midsagittal alignment have been implemented | ||
<!-- Fill this out before Friday's summary presentations - list what you did and how well it worked. --> | <!-- Fill this out before Friday's summary presentations - list what you did and how well it worked. --> | ||
| − | |||
</div> | </div> | ||
| Line 44: | Line 50: | ||
==References== | ==References== | ||
| − | + | The fiber laterality histogram: A new way to measure white matter asymmetry. | |
| + | Lauren J. O'Donnell, Carl-Fredrik Westin, Isaiah Norton, Stephen Whalen, Laura Rigolo, Ruth Propper, and Alexandra J. Golby | ||
| + | Med Image Comput Comput Assist Interv. 2010; 13(Pt 2): 225-232 | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
Latest revision as of 14:58, 24 June 2011
Home < 2011 Summer Project Week White Matter LateralityWhite Matter Laterality/Asymmetry Measurement
Key Investigators
- Harvard Medical School: Lauren O'Donnell
- Kitware: JC (Thanks for help on how to integrate as a Slicer4 module/extension)
- Harvard Medical School: Sandy Wells (Thanks for helpful discussions on angle representation and objective functions for registration)
Objective
- Fast computation of white matter
- fiber tract laterality indices
- in python and ... Slicer4
Approach, Plan
- So far have preliminary code running in python/vtk
- Slicer-ification is goal for this week
- Another goal: learn how to make an extension module
- Another goal: can this module use testing framework
Progress
- Project is now organized as a python package
- Command line script written to run on large groups
- Initial testing on R/L handedness group data showed significant differences
- Preprocessing and midsagittal alignment have been implemented
References
The fiber laterality histogram: A new way to measure white matter asymmetry. Lauren J. O'Donnell, Carl-Fredrik Westin, Isaiah Norton, Stephen Whalen, Laura Rigolo, Ruth Propper, and Alexandra J. Golby Med Image Comput Comput Assist Interv. 2010; 13(Pt 2): 225-232
Delivery Mechanism
This work will be delivered to the NAMIC Kit as a
- NITRIC distribution
- Slicer Module
- Built-in:
- Extension -- commandline:
- Extension -- loadable: