Difference between revisions of "Projects:InteractiveSegmentation"
Ivan.kolesov (talk | contribs) (Created page with ' Back to Georgia Tech Algorithms __NOTOC__ = Adaptive Radiotherapy for head, neck and thorax = Proton therapy is used to deliver accurate doses of radiation…') |
|||
| (13 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[Algorithm: | + | Back to [[Algorithm:Stony Brook|Stony Brook University Algorithms]] |
__NOTOC__ | __NOTOC__ | ||
| − | = | + | = Interactive Image Segmentation With Active Contours = |
| − | + | A driving clinical study for the present work is a population study of skeletal development in youth. Bone grows from the physis (growth plate), located in the middle of a long bone between the epiphysis and metaphysis. Full adult growth is reached when the physis disappears completely. Precise understanding of how the growth plates in femur and tibia change from childhood to adulthood enables improved surgical planning(e.g. determining tunnel placement for anterior cruciate ligament (ACL) reconstruction) to avoid stunting the patient's growth or compromising the stability of the knee. Currently, growth potential is measured by a physician using an x-ray scan of multiple bones; patients receive repeated doses of radiation exposure. | |
= Description = | = Description = | ||
| + | In this work, interactive segmentation is integrated with an active contour model and segmentation is posed as a human-supervisory control problem. User input is tightly coupled with an automatic segmentation algorithm leveraging the user's high-level anatomical knowledge and the automated method's speed. Real-time visualization enables the user to quickly identify and correct the result in a sub-domain where the variational model's statistical assumptions do not agree with his expert knowledge. Methods developed in this work are applied to magnetic resonance imaging (MRI) volumes as part of a population study of human skeletal development. Segmentation time is reduced by approximately five times over similarly accurate manual segmentation of large bone structures. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | * [[Image:KSliceFlowChart.png | Rel Pred| 800px]] | ||
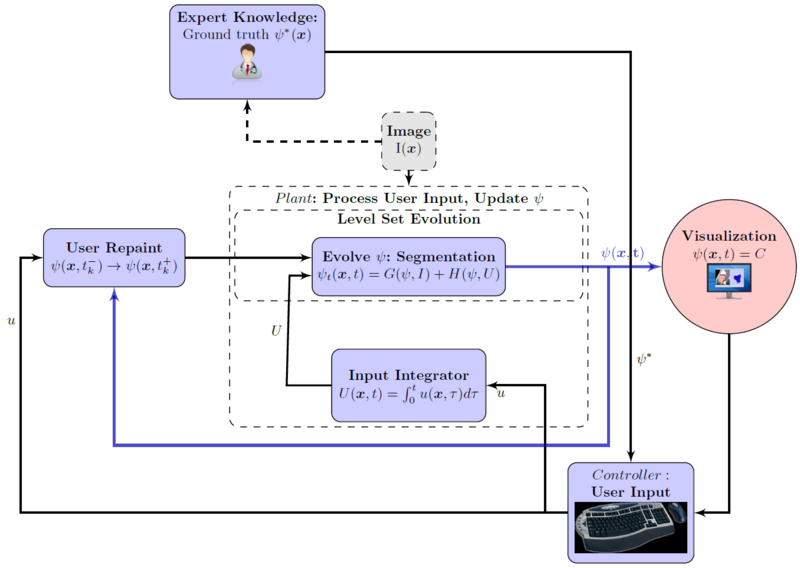
| + | Flowchart for the interactive segmentation approach. Notice the user's pivotal role in the process. | ||
| + | * [[Image:KSliceInptTimeChart.png | Eye Seg| 800px]] | ||
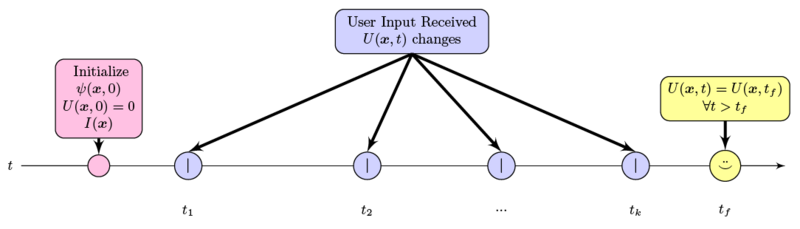
| + | Time-line of user input into the system. Note that user input is sparse, has local effect only, and decreases in frequency and magnitude over time. | ||
| + | * [[Image:KVoutSegTightMod.png | Eye Seg| 300px]] | ||
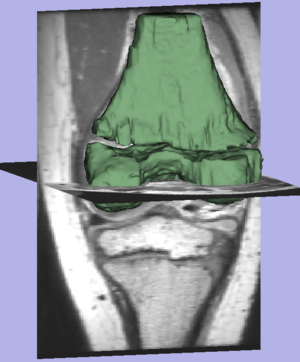
| + | Result of the segmentation. | ||
== Current State of Work == | == Current State of Work == | ||
| − | We have | + | The described algorithm is implemented in c++ and delivered to physicians. We have begun to analyze the data they created by segmenting the knee with out tool. Future work incorporates shape prior into the segmentation and improves user interaction(according to feedback physician's provide us). |
| + | = Key Investigators = | ||
| − | |||
| − | * Georgia Tech: Ivan Kolesov, | + | * Georgia Tech: Ivan Kolesov, Peter Karasev, and Karol Chudy |
| − | * | + | * Boston University: Allen Tannenbaum |
| + | * Emory University: Grant Muller and John Xerogeanes | ||
= Publications = | = Publications = | ||
| Line 26: | Line 30: | ||
''In Press'' | ''In Press'' | ||
| − | I. Kolesov, | + | I. Kolesov, P.Karasev, G.Muller, K.Chudy, J.Xerogeanes, and A. Tannenbaum. Human Supervisory Control Framework for Interactive Medical Image Segmentation. MICCAI Workshop on Computational Biomechanics for Medicine 2011. |
| + | |||
| + | |||
| + | P.Karasev, I.Kolesov, K.Chudy, G.Muller, J.Xerogeanes, and A. Tannenbaum. Interactive MRI Segmentation with Controlled Active Vision. IEEE CDC-ECC 2011. | ||
Latest revision as of 01:03, 16 November 2013
Home < Projects:InteractiveSegmentationBack to Stony Brook University Algorithms
Interactive Image Segmentation With Active Contours
A driving clinical study for the present work is a population study of skeletal development in youth. Bone grows from the physis (growth plate), located in the middle of a long bone between the epiphysis and metaphysis. Full adult growth is reached when the physis disappears completely. Precise understanding of how the growth plates in femur and tibia change from childhood to adulthood enables improved surgical planning(e.g. determining tunnel placement for anterior cruciate ligament (ACL) reconstruction) to avoid stunting the patient's growth or compromising the stability of the knee. Currently, growth potential is measured by a physician using an x-ray scan of multiple bones; patients receive repeated doses of radiation exposure.
Description
In this work, interactive segmentation is integrated with an active contour model and segmentation is posed as a human-supervisory control problem. User input is tightly coupled with an automatic segmentation algorithm leveraging the user's high-level anatomical knowledge and the automated method's speed. Real-time visualization enables the user to quickly identify and correct the result in a sub-domain where the variational model's statistical assumptions do not agree with his expert knowledge. Methods developed in this work are applied to magnetic resonance imaging (MRI) volumes as part of a population study of human skeletal development. Segmentation time is reduced by approximately five times over similarly accurate manual segmentation of large bone structures.
Flowchart for the interactive segmentation approach. Notice the user's pivotal role in the process.
Time-line of user input into the system. Note that user input is sparse, has local effect only, and decreases in frequency and magnitude over time.
Result of the segmentation.
Current State of Work
The described algorithm is implemented in c++ and delivered to physicians. We have begun to analyze the data they created by segmenting the knee with out tool. Future work incorporates shape prior into the segmentation and improves user interaction(according to feedback physician's provide us).
Key Investigators
- Georgia Tech: Ivan Kolesov, Peter Karasev, and Karol Chudy
- Boston University: Allen Tannenbaum
- Emory University: Grant Muller and John Xerogeanes
Publications
In Press
I. Kolesov, P.Karasev, G.Muller, K.Chudy, J.Xerogeanes, and A. Tannenbaum. Human Supervisory Control Framework for Interactive Medical Image Segmentation. MICCAI Workshop on Computational Biomechanics for Medicine 2011.
P.Karasev, I.Kolesov, K.Chudy, G.Muller, J.Xerogeanes, and A. Tannenbaum. Interactive MRI Segmentation with Controlled Active Vision. IEEE CDC-ECC 2011.