Difference between revisions of "2013 Project Week:WMH Segmentation for Stroke"
From NAMIC Wiki
| (3 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
Image:PW-SLC2013.png|[[2013_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2013.png|[[2013_Winter_Project_Week#Projects|Projects List]] | ||
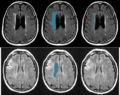
Image:WMH_T1.png| T1 images in stroke dataset. | Image:WMH_T1.png| T1 images in stroke dataset. | ||
| − | Image:WMHseg.png | left: FLAIR images, middle: manual delineation of | + | Image:WMHseg.png | left: FLAIR images, middle: manual delineation of relevant areas, right: manual WMH segmentation. |
</gallery> | </gallery> | ||
| Line 17: | Line 17: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | We are developing methods for segmentation of white matter hyperintensity in FLAIR images of stroke patients. Specifically, WMH is to be localized in particular areas of the brain, as seen in a training set. This dataset is particularly challenging due to the low resolution (1mm x 1mm x 7mm) and with cropped fields of view in the given images. | + | We are developing methods for segmentation of white matter hyperintensity (WMH) in FLAIR images of stroke patients. Specifically, WMH is to be localized in particular areas of the brain, as seen in a training set. This dataset is particularly challenging due to the low resolution (1mm x 1mm x 7mm) and with cropped fields of view in the given images. |
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| Line 29: | Line 29: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | * Aside from a in-house model for cropped field of view registration we also tuned ANTS to our specific images and pre-processing |
| + | * We wrote a pipeline to parse through the nearly one thousand subjects | ||
| + | * We built several atlases based on the Buckner40 dataset using ANTS | ||
| + | * We wrote a pipeline to register T1 scans to an atlas, and FLAIR images to T1 images | ||
| + | * We wrote and tested some classification methods using SVMs with RBF kernels based on intensity features. We currently don't impose a smoothness constraint, but can optionally run a slight smoothness pre-processing step. | ||
| + | * Whole pipeline is nearly done, currently dealing with slight bugs :) | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 22:59, 10 January 2013
Home < 2013 Project Week:WMH Segmentation for Stroke
Key Investigators
- Adrian Dalca, Ramesh Sridharan, Polina Golland, MIT
- Natalia Rost, Jonathan Rosand, MGH
Project Description
Objective
We are developing methods for segmentation of white matter hyperintensity (WMH) in FLAIR images of stroke patients. Specifically, WMH is to be localized in particular areas of the brain, as seen in a training set. This dataset is particularly challenging due to the low resolution (1mm x 1mm x 7mm) and with cropped fields of view in the given images.
Approach, Plan
- Find and apply appropriate registration method to register cropped, low resolution T1 scans
- Register T1 scans to FLAIR images with rigid registration
- Create mask of relevant areas for WMH segmentation from manual segmentations
- Investigate several known methods for segmentation of WMH segmentation using training images.
- Write thorough pipline for above steps.
Progress
- Aside from a in-house model for cropped field of view registration we also tuned ANTS to our specific images and pre-processing
- We wrote a pipeline to parse through the nearly one thousand subjects
- We built several atlases based on the Buckner40 dataset using ANTS
- We wrote a pipeline to register T1 scans to an atlas, and FLAIR images to T1 images
- We wrote and tested some classification methods using SVMs with RBF kernels based on intensity features. We currently don't impose a smoothness constraint, but can optionally run a slight smoothness pre-processing step.
- Whole pipeline is nearly done, currently dealing with slight bugs :)