Difference between revisions of "2013 Summer Project Week:TractAtlasCluster"
(Created page with '__NOTOC__ <gallery> Image:PW-MIT2013.png|Projects List Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus…') |
|||
| (9 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2013.png|[[2013_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2013.png|[[2013_Summer_Project_Week#Projects|Projects List]] | ||
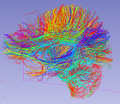
| − | Image: | + | Image:Clusters.png|Tract clusters in a healthy control brain. |
| − | Image: | + | Image:Clusters_points.png|Clusters across subjects, relative to landmarks from fMRI. |
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
* BWH: Lauren O'Donnell | * BWH: Lauren O'Donnell | ||
| − | * | + | * Slicer gurus: thanks Steve Pieper |
| + | * Open source/python/vtk gurus: thanks Bill Lorensen | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 21: | Line 15: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | The goal is to work on Slicer integration and user packaging | + | The goal is to work on Slicer integration and user packaging to enable open-source release of Python tract atlasing and clustering code. |
| Line 31: | Line 25: | ||
Our python package includes code implementing tract clustering, atlas labeling, registration, and fMRI-DTI modeling as part of the atlas. References are below. | Our python package includes code implementing tract clustering, atlas labeling, registration, and fMRI-DTI modeling as part of the atlas. References are below. | ||
| − | Our plan for the project week is to focus on Slicer compatibility and user accessibility of the code. | + | Our plan for the project week is to focus on Slicer compatibility and user accessibility/cleanup/documentation of the code. |
</div> | </div> | ||
| Line 38: | Line 32: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | Provided source code to collaborators in Toronto, and code was installed and tested successfully at their lab. New test scripts were written. Code was updated and initially tested to work with vtk6. For possible ease of distribution, code was tested against a new install of Canopy (new Enthought python) distribution (that includes scipy) with their provided vtk package. However that vtk package lacked some vtkCellArray wrappings needed by our code (that are present when compiling and wrapping vtk from source). A bug report was filed with Enthought regarding the wrappings, and a discussion with Bill Lorensen indicated it is due to an older version of vtk distributed with Canopy. Enthought has now told us they will update their vtk package to a more recent vtk5. Other progress: Discussions with Steve Pieper about likely/planned python packages in slicer. | |
</div> | </div> | ||
| Line 52: | Line 46: | ||
##Extension -- commandline | ##Extension -- commandline | ||
##Extension -- loadable | ##Extension -- loadable | ||
| − | #Other (Please specify) | + | #Other (Please specify) YES: eventual plan is an extension but depends on packages not yet in Slicer python. |
==References== | ==References== | ||
| + | * Fiber clustering versus the parcellation-based connectome | ||
| + | LJ O’Donnell, AJ Golby, CF Westin | ||
| + | NeuroImage 2013 | ||
* Unbiased Groupwise Registration of White Matter Tractography | * Unbiased Groupwise Registration of White Matter Tractography | ||
LJ O’Donnell, WM Wells III, AJ Golby, CF Westin | LJ O’Donnell, WM Wells III, AJ Golby, CF Westin | ||
Latest revision as of 14:12, 21 June 2013
Home < 2013 Summer Project Week:TractAtlasClusterKey Investigators
- BWH: Lauren O'Donnell
- Slicer gurus: thanks Steve Pieper
- Open source/python/vtk gurus: thanks Bill Lorensen
Objective
The goal is to work on Slicer integration and user packaging to enable open-source release of Python tract atlasing and clustering code.
Approach, Plan
Our python package includes code implementing tract clustering, atlas labeling, registration, and fMRI-DTI modeling as part of the atlas. References are below. Our plan for the project week is to focus on Slicer compatibility and user accessibility/cleanup/documentation of the code.
Progress
Provided source code to collaborators in Toronto, and code was installed and tested successfully at their lab. New test scripts were written. Code was updated and initially tested to work with vtk6. For possible ease of distribution, code was tested against a new install of Canopy (new Enthought python) distribution (that includes scipy) with their provided vtk package. However that vtk package lacked some vtkCellArray wrappings needed by our code (that are present when compiling and wrapping vtk from source). A bug report was filed with Enthought regarding the wrappings, and a discussion with Bill Lorensen indicated it is due to an older version of vtk distributed with Canopy. Enthought has now told us they will update their vtk package to a more recent vtk5. Other progress: Discussions with Steve Pieper about likely/planned python packages in slicer.
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below)
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline
- Extension -- loadable
- Other (Please specify) YES: eventual plan is an extension but depends on packages not yet in Slicer python.
References
- Fiber clustering versus the parcellation-based connectome
LJ O’Donnell, AJ Golby, CF Westin NeuroImage 2013
- Unbiased Groupwise Registration of White Matter Tractography
LJ O’Donnell, WM Wells III, AJ Golby, CF Westin Medical Image Computing and Computer-Assisted Intervention–MICCAI 2012, 123-130
- fMRI-DTI modeling via landmark distance atlases for prediction and detection of fiber tracts
LJ O'Donnell, L Rigolo, I Norton, WM Wells III, CF Westin, AJ Golby NeuroImage 60 (1), 456-70
- Automatic tractography segmentation using a high-dimensional white matter atlas
LJ O'Donnell, CF Westin Medical Imaging, IEEE Transactions on 26 (11), 1562-1575