Difference between revisions of "2014 How about the Future"
From NAMIC Wiki
| (31 intermediate revisions by 5 users not shown) | |||
| Line 12: | Line 12: | ||
This page contains talking points for the opening session of the NA-MIC AHM 2014 (the last one). | This page contains talking points for the opening session of the NA-MIC AHM 2014 (the last one). | ||
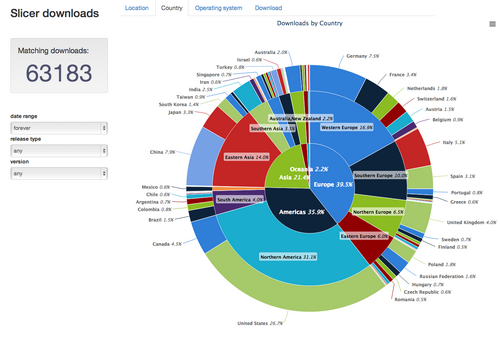
| − | | style="background: #ebeced | + | | style="background: #ebeced" align="center"| [[Image:Slicer-country-stats-2013-11-16.png|500px|Slicer downloads by country and region]] |
| + | Slicer downloads by country and region | ||
|- | |- | ||
|} | |} | ||
| Line 18: | Line 19: | ||
=What we have accomplished= | =What we have accomplished= | ||
| − | *Created | + | *Created a vibrant scientific and engineering community in the field of [http://en.wikipedia.org/wiki/Medical_image_computing Medical Image Computing] (MIC). |
| − | *Investigated novel algorithmic approaches: Particle systems, registration algorithms, segmentation algorithms, | + | *Investigated novel algorithmic approaches: Particle systems, registration algorithms, segmentation algorithms, software engineering methodologies |
*Created the NA-MIC kit, a free open source platform for MIC and the basis for 3D Slicer. | *Created the NA-MIC kit, a free open source platform for MIC and the basis for 3D Slicer. | ||
*3D Slicer is today a platform with worldwide impact. | *3D Slicer is today a platform with worldwide impact. | ||
| Line 25: | Line 26: | ||
=Highlights= | =Highlights= | ||
* Robust algorithms for segmentation in the face of anatomical variability: label fusion | * Robust algorithms for segmentation in the face of anatomical variability: label fusion | ||
| + | * Comprehensive toolbox for the analysis of individual and population DTI data | ||
| + | * The predominant shape analysis toolbox in the field/used in clinical papers(SPHARM-PDM) | ||
* A novel framework for modeling brain connectivity networks | * A novel framework for modeling brain connectivity networks | ||
* Robust pipeline for processing clinical brain images | * Robust pipeline for processing clinical brain images | ||
| + | * Automatic 4D segmentation of longitudinal brain MRI in severe TBI | ||
| + | * Tract-oriented white matter analysis for subject-specific and population analysis | ||
| + | * NAMIC researchers take lead in novel spatio-temporal image and shape analysis methodologies (Whitaker, Styner, Gerig, Tannenbaum and colleagues) | ||
=Where we go from here= | =Where we go from here= | ||
| − | *Project week will continue. | + | *Project week will continue to provide a venue for hands on multi-partner collaboration in a rich MIC software environment. |
| − | *3D Slicer 4 | + | *3D Slicer 4 will continue to provide a stable platform as a target for plug-in development and distribution. |
| − | *The remaining months of NA-MIC funding will be used to simplify the submission of extensions. | + | *The remaining months of NA-MIC funding will be used to |
| − | * | + | **Improve the stability of Slicer and simplify the submission of extensions. |
| − | **NIH mandated sunset for NA-MIC in June 2014. | + | **Create a new batch of extensions |
| − | **[http://projectreporter.nih.gov/project_info_description.cfm?aid=8415024&icde=18711051&ddparam=&ddvalue=&ddsub=&cr=8&csb=default&cs=ASC NAC] funded through 2018. | + | **Improve documentation and create tutorials |
| − | **[http://projectreporter.nih.gov/project_info_description.cfm?aid=8606944&icde=18711051&ddparam=&ddvalue=&ddsub=&cr=1&csb=default&cs=ASC QIICR] funded through 2018. | + | |
| + | =Grants= | ||
| + | *Grants that will allow continued participation in the community after the NIH mandated sunset for NA-MIC in June 2014. | ||
| + | **[http://projectreporter.nih.gov/project_info_description.cfm?aid=8415024&icde=18711051&ddparam=&ddvalue=&ddsub=&cr=8&csb=default&cs=ASC NAC] funded through 2018. Ron Kikinis | ||
| + | **[http://projectreporter.nih.gov/project_info_description.cfm?aid=8606944&icde=18711051&ddparam=&ddvalue=&ddsub=&cr=1&csb=default&cs=ASC QIICR] funded through 2018. Ron Kikinis, Andrey Fedorov | ||
| + | ** "An Open Source Software for Proton Treatment Planning," NCI/Federal Share Grant C06-CA059267 funded through Dec 2014, Greg Sharp | ||
| + | **[http://projectreporter.nih.gov/project_info_description.cfm?aid=8551816&icde=18802965&ddparam=&ddvalue=&ddsub=&cr=1&csb=default&cs=ASC 4DShape]"4D Shape Analysis for Modeling Spatiotemporal Change Trajectories in Huntington's" (NINDS, 07/01/12- 06/30/15) PIs Gerig/Fletcher Utah, co-investigators Johnson/Paulsen Iowa | ||
| + | ** [http://projectreporter.nih.gov/project_info_description.cfm?aid=8576556&icde=18804072 NA-MIC Collaboration R01] (NIDCR) on "Quantification of 3D Bony Changes in Temporomandibular joint osteoarthritis", PI Cevidanes/U Michigan, co-investigators Martin Styner/Beatriz Paniagua/UNC, Steve Piper | ||
**Several other grants are being worked on | **Several other grants are being worked on | ||
| − | + | =Software= | |
| − | + | *3D Slicer | |
| − | * " | + | *Other packages and Slicer extensions |
| − | * " | + | ** "MABS," an end-user software for multi-atlas segmentation, Greg Sharp |
| + | ** "SPHARM-PDM", an end-user software for shape analysis, also including group-wise based particle correspondence, Martin Styner | ||
| + | ** "DTIPrep", Comprehensive DWI/DTI QC tool, Martin Styner | ||
| + | ** "DTIAtlasBuilder", DTI co-registration and atlas building software, Martin Styner | ||
| + | ** "DTIAtlasFiberAnalyzer", DTI fiber profile statistics generation, Martin Styner | ||
| + | ** "FiberViewerLight", DTI fiber processing & clustering software, Martin Styner | ||
| + | ** "CARMA Cardiac MR Tookit", Cardiac MRI segmentation and quantification software, CARMA Center | ||
| + | ** "ABC: Atlas-based Classification", Automatic brain MRI processing pipeline, Prastawa/Gerig | ||
| + | ** "SlicerRT", adaptive radiation therapy data analysis, comparison, and visualization tools, Gabor Fichtinger | ||
| + | ** "SlicerIGT", image-guided therapy planning and guidance tools, Nobuhiko Hata, Gabor Fichtinger | ||
| + | ** "PerkTutor", training and skill assessment software for needle-based percutaneous interventions, Gabor Fichtinger | ||
| + | ** "MatlabBridge", tool for making available software developed in Matlab environment inside 3D Slicer, Gabor Fichtinger | ||
| + | ** "TransformVisualizer", software for quick visual assessment of registration results, Gabor Fichtinger | ||
Latest revision as of 13:50, 9 January 2014
Home < 2014 How about the FutureBack to AHM_2014 Agenda
What we have accomplished
- Created a vibrant scientific and engineering community in the field of Medical Image Computing (MIC).
- Investigated novel algorithmic approaches: Particle systems, registration algorithms, segmentation algorithms, software engineering methodologies
- Created the NA-MIC kit, a free open source platform for MIC and the basis for 3D Slicer.
- 3D Slicer is today a platform with worldwide impact.
Highlights
- Robust algorithms for segmentation in the face of anatomical variability: label fusion
- Comprehensive toolbox for the analysis of individual and population DTI data
- The predominant shape analysis toolbox in the field/used in clinical papers(SPHARM-PDM)
- A novel framework for modeling brain connectivity networks
- Robust pipeline for processing clinical brain images
- Automatic 4D segmentation of longitudinal brain MRI in severe TBI
- Tract-oriented white matter analysis for subject-specific and population analysis
- NAMIC researchers take lead in novel spatio-temporal image and shape analysis methodologies (Whitaker, Styner, Gerig, Tannenbaum and colleagues)
Where we go from here
- Project week will continue to provide a venue for hands on multi-partner collaboration in a rich MIC software environment.
- 3D Slicer 4 will continue to provide a stable platform as a target for plug-in development and distribution.
- The remaining months of NA-MIC funding will be used to
- Improve the stability of Slicer and simplify the submission of extensions.
- Create a new batch of extensions
- Improve documentation and create tutorials
Grants
- Grants that will allow continued participation in the community after the NIH mandated sunset for NA-MIC in June 2014.
- NAC funded through 2018. Ron Kikinis
- QIICR funded through 2018. Ron Kikinis, Andrey Fedorov
- "An Open Source Software for Proton Treatment Planning," NCI/Federal Share Grant C06-CA059267 funded through Dec 2014, Greg Sharp
- 4DShape"4D Shape Analysis for Modeling Spatiotemporal Change Trajectories in Huntington's" (NINDS, 07/01/12- 06/30/15) PIs Gerig/Fletcher Utah, co-investigators Johnson/Paulsen Iowa
- NA-MIC Collaboration R01 (NIDCR) on "Quantification of 3D Bony Changes in Temporomandibular joint osteoarthritis", PI Cevidanes/U Michigan, co-investigators Martin Styner/Beatriz Paniagua/UNC, Steve Piper
- Several other grants are being worked on
Software
- 3D Slicer
- Other packages and Slicer extensions
- "MABS," an end-user software for multi-atlas segmentation, Greg Sharp
- "SPHARM-PDM", an end-user software for shape analysis, also including group-wise based particle correspondence, Martin Styner
- "DTIPrep", Comprehensive DWI/DTI QC tool, Martin Styner
- "DTIAtlasBuilder", DTI co-registration and atlas building software, Martin Styner
- "DTIAtlasFiberAnalyzer", DTI fiber profile statistics generation, Martin Styner
- "FiberViewerLight", DTI fiber processing & clustering software, Martin Styner
- "CARMA Cardiac MR Tookit", Cardiac MRI segmentation and quantification software, CARMA Center
- "ABC: Atlas-based Classification", Automatic brain MRI processing pipeline, Prastawa/Gerig
- "SlicerRT", adaptive radiation therapy data analysis, comparison, and visualization tools, Gabor Fichtinger
- "SlicerIGT", image-guided therapy planning and guidance tools, Nobuhiko Hata, Gabor Fichtinger
- "PerkTutor", training and skill assessment software for needle-based percutaneous interventions, Gabor Fichtinger
- "MatlabBridge", tool for making available software developed in Matlab environment inside 3D Slicer, Gabor Fichtinger
- "TransformVisualizer", software for quick visual assessment of registration results, Gabor Fichtinger