Difference between revisions of "2014 Project Week:TBISegmentation"
From NAMIC Wiki
| (3 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-SLC2014.png|[[2014_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2014.png|[[2014_Winter_Project_Week#Projects|Projects List]] | ||
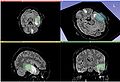
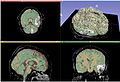
| − | Image: | + | Image:AcSeg_initial_box.jpg| Initial bounding box |
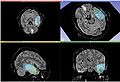
| + | Image:AcSeg_selftraining.jpg| Self training update | ||
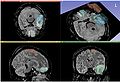
| + | Image:AcSeg_activelearning.jpg| Active learning update | ||
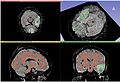
| + | Image:AcSeg_final_alphamap.jpg| Final alpha map | ||
| + | Image:AcSeg_final_seg_normal_tissue_classes.jpg| Segmentation of normal tissue classes | ||
</gallery> | </gallery> | ||
| + | |||
==Key Investigators== | ==Key Investigators== | ||
| Line 32: | Line 37: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | * Ran the command line tool with Andrei step by step to show how it works and test it. |
| + | * Meeting with Jack and Andrei to discuss sharing the current command line tool with them. | ||
| + | * Joined the TBI DBP meeting to discuss how to integrate the current command line tool into Slicer 3D. | ||
| + | ** Stephen and Steve recommended to do a Python scripting module and talk to Jean-Christophe. | ||
| + | ** Talked to Jean-Christophe and he pointed me a tutorial [http://www.na-mic.org/Wiki/index.php/2013_Project_Week_Breakout_Session:Slicer4Python the wiki of breakout session Slicer4Python]. | ||
| + | ** Tried to understand the Slicer4Python tutorial and followed it step by step (still working on). | ||
| + | ** Talked to other Slicer developers to ask for help and suggestions. | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 06:12, 10 January 2014
Home < 2014 Project Week:TBISegmentation
Key Investigators
- GE: Marcel Prastawa
- Utah: Bo Wang, Guido Gerig
- USC: Andrei Irimia, Jack Van Horn
Project Description
Objective
- Develop a pipeline combining our TBI segmentation algorithm with other Slicer modules.
- Study and gain understanding of the manual process for segmenting TBI.
- Test the algorithm with users from USC.
Approach, Plan
- Wrap existing code into a Slicer extension.
- Determine approaches for user interaction for semi-automatic segmentation.
- Test and evaluate Slicer modules for TBI processing:
- Intra and inter time points coregistration.
- Initial atlas alignment using affine transformation.
- Skull stripping to obtain initial brain mask.
Progress
- Ran the command line tool with Andrei step by step to show how it works and test it.
- Meeting with Jack and Andrei to discuss sharing the current command line tool with them.
- Joined the TBI DBP meeting to discuss how to integrate the current command line tool into Slicer 3D.
- Stephen and Steve recommended to do a Python scripting module and talk to Jean-Christophe.
- Talked to Jean-Christophe and he pointed me a tutorial the wiki of breakout session Slicer4Python.
- Tried to understand the Slicer4Python tutorial and followed it step by step (still working on).
- Talked to other Slicer developers to ask for help and suggestions.